Back to Journals » Infection and Drug Resistance » Volume 16
Phenotypic Drug Resistance Pattern and Mutation Characteristics of Mycobacterium tuberculosis from Different Body Fluids Among Extra Pulmonary Patients Presented in Selected Hospitals in Addis Ababa, Ethiopia
Authors Alehegn E, Gebreyohanns A, Berhane BW, Wright JA, Hundie GB, Geremew RA, Gorems K , Gebreyohannes Z , Amare M, Abebaw Y, Diriba G , Zerihun B, Gebremichael AW, Kassa M, Gize A
Received 6 April 2023
Accepted for publication 11 August 2023
Published 22 August 2023 Volume 2023:16 Pages 5511—5522
DOI https://doi.org/10.2147/IDR.S415906
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Elias Alehegn,1,2 Alganesh Gebreyohanns,1 Bereket Woldeab Berhane,3 Jayne A Wright,4 Gadissa Bedada Hundie,1 Rozina Ambachew Geremew,1 Kasahun Gorems,1 Zenebe Gebreyohannes,1 Misikir Amare,5 Yeshiwork Abebaw,5 Getu Diriba,5 Betselot Zerihun,5 Amanuel Wondimu Gebremichael,5 Melkayehu Kassa,1 Addisu Gize1,6
1Department of Microbiology, St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia; 2Addis Ababa City Administration, Farmers and Urban Agriculture Development Commission, Addis Ababa, Ethiopia; 3Department of Pathology, St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia; 4Jayne Wright Ltd, Pathology Consultancy, Ledbury, UK; 5Ethiopian Public Health Institute, Addis Ababa, Ethiopia; 6CIHLMU Center for International Health, LMU University Hospital, Munich, Germany
Correspondence: Elias Alehegn, Department of Microbiology, St. Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia, Email [email protected]
Background: Drug resistance in tuberculosis poses challenges to both the control and prevention of the disease. The extent of resistance is not well known in developing countries, including Ethiopia. This study was conducted to determine the drug resistance patterns and mutation characteristics of Mycobacterium tuberculosis among extra pulmonary tuberculosis patients in selected health facilities in Addis Ababa.
Material and Methods: A cross-sectional study was conducted from February 2022 to August 2022 in selected hospitals in Addis Ababa. Socio-demographic and clinical data were collected using structured questionnaire. Mycobacterium tuberculosis complex (MTBC) isolates were tested for phenotypic drug susceptibility patterns using the Mycobacterium growth indicator tube (MGIT) method for first-line drugs and mutation characteristics using the Line Probe Assay (LPA) method. The data were analyzed using: SPSS version 23, and a P-value ≤ 0.05 was considered statistically significant.
Results: From a total of 308 patient samples from presumptive extra pulmonary patients, 44 (14.3%) were positive for MTBC. Any drug resistance was discovered in 25% of 44 MTBC isolates evaluated for five first-line drugs phenotypically, with isoniazid (INH) and pyrazinamide (PZA) resistance accounting for a greater proportion with 13.6% and 11.4% of the isolates, respectively. Two (4.5%) of the isolates were MDR-TB. Out of 44 isolates tested using the Geno Type MTBDRplus assay, 5 (11.4%) showed mutations at katG and 2 (4.5%) showed mutations in the rpoB genes.
Conclusion: Both the phenotypic and genotypic drug susceptibility test results showed a high proportion of INH resistance. All INH resistance-conferring mutations were identified from katG gene. The overall prevalence of MDR-TB was also high. For early case detection and treatment, expanding diagnostic capacity for first-line DST is a vital step to limit further spread of drug resistant TB strains in the study area.
Keywords: extra pulmonary tuberculosis, Mycobacterium tuberculosis, drug resistance
Introduction
Antimicrobial resistance (AMR) is becoming a global public health threat. The problem, which is being exacerbated by antibiotic misuse, is directly or indirectly causing severe and complicated infections, longer periods of hospitalization, and higher mortality rates. Specifically, antibiotic overuse aggravates side effects, increasing the need for medication for even normally self-limiting diseases.1 Resistance to anti-TB drugs is caused by different clinical, biological, and microbiological factors. Non-adherence of patients to treatment, errors by physicians in the management of disease treatment, and problems with drugs reaching the target site as a result of poor vascularization caused by granulomatous lesions are some of the host-related factors. Intrinsic drug resistance, non-replicating drug-tolerant bacilli residing within granulomas, and mutations in bacterial genes are pathogen-related factors.2
In 2021, there are expected to be 450,000 incident cases of MDR/RR-TB worldwide, up 3.1% from 437,000 in 2020. The major reason for this rise is an overall increase in TB incidence between 2020 and 2021, which is thought to have happened as a result of the COVID-19 pandemic’s influence on TB detection. In 2021, an expected 191000 people died as a result of MDR/RR-TB. The predicted proportion of new MDR/RR-TB infections was 3.9% in 2015 and 3.6% in 2021, with previously treated cases accounting for 20% in 2015 and 18% in 2021. In Ethiopia, the incidence of MDR/RR TB patients was 1800 in 2021, with 1.1% new cases and 12% previously treated cases.3
Multi-drug resistant TB remains a clear obstacle to the control and eradication of TB. There is a lack of data on the incidence of drug-resistant TB in many countries, particularly in sub-Saharan Africa.4 Multi-drug resistant TB emerges primarily from poor management of those affected by the disease, which requires appropriate diagnosis and timely treatment in order for effective containment. Thus, efforts must be focused on the correct medications in each new case.5 In recent years, reports show that the global burden of MDR-TB has risen by more than 20%, and only half of all MDR-TB patients are being treated successfully, according to the WHO report.6 The proportion of drug-resistant extra pulmonary TB (EPTB) is also increasing, but unfortunately, very little data on the prevalence and impact of drug-resistant TB are available in the medical literature.7 This may be because of the diagnostic challenges of EPTB as a result of the diverse presenting signs of the disease.8
Although MDR and XDR tuberculosis are present worldwide, drug susceptibility testing (DST) is mainly performed when the requisite resources are available rather than as a routine to monitor drug-resistance rates. Phenotypic methods of DST are widely used in developed countries, whereas in developing countries genotypic methods are used to avoid culture and biosafety constraints. Genotypic methods show only the screen for mutations in resistance genes, while phenotypic methods detect resistance accompanied by risk factors associated with the problem.9 There is a scarcity of data on the drug-specific resistance patterns of EPTB, particularly in countries with high TB burdens, as a result of the lack of extra pulmonary specimens and drug susceptibility testing capabilities.10 Unlike pulmonary TB, systematic drug resistance surveillance in EPTB is not carried out, and consequently there are no reliable estimates of the level of drug resistance in EPTB.7 Hence, the primary objective of this study was to determine the drug resistance patterns and mutation characteristics of Mycobacterium tuberculosis isolates obtained from extra pulmonary tuberculosis patients in selected health facilities in Addis Ababa, Ethiopia.
Materials and Methods
Study Setting
A cross-sectional study was conducted from February 2022 to August 2022 to determine and assess the phenotypic drug resistance pattern and mutation characteristics of M. tuberculosis isolates obtained from extra pulmonary patients presented in referral and non-referral selected hospitals found in Addis Ababa, Ethiopia. A total of three public hospitals were selected. The selected hospitals were St. Paul’s Hospital Millennium Medical College, Ras Desta Damtew Memorial Hospital, and Zewditu Memorial Hospitals.
Study Population and Sample Size
The source of the study population was those patients who visited the selected hospitals for medical services. The sample size was estimated from the previous study reported in the study area to determine the prevalence of extra pulmonary tuberculosis among presumptive patients. Confirmed extra pulmonary patients were obtained from laboratory tested presumptive extra pulmonary patients. The study population confirmed extra pulmonary tuberculosis patients. Age, sex, marital status, educational status, residency, aspirate type, EPTB case type, and contact with known TB cases were independent variables. The study participants were contacted at wards during their clinical examination. Those who were diagnosed as presumptive EPTB patients and met the inclusion criteria were considered for sample collection after they gave written consent.
Inclusion and Exclusion Criteria
All culture-positive isolates presented with a correct patient identification number and having demographic data were included in this study. Those patients who were actively taking anti-TB drug during the study period were excluded from the study.
Laboratory Methods
Sample Collection and Mycobacterial Isolation
Cerebrospinal fluid and other body fluids were collected using a fine needle aspirate technique collection. The collected clinical samples were taken to the National Tuberculosis Reference Laboratory (NTRL) via a cold chain. The laboratory is repeatedly accredited by different organizations and found at the Ethiopian Public Health Institute. Samples were processed immediately and stored at 4°C if there was any delay to be processed.
Both BACTEC MGIT 960 (BBL® MGIT™ System) and LJ (egg-based) medium culture techniques were used to isolate bacteria. Samples were digested using freshly prepared N-acetyl-L-cysteine (NALC) and decontaminated by NaOH (1%). A 50 mL of NaOH-Na citrate solution was added to 0.25 g NALC powder to achieve a final concentration of 0.5%. Phosphate buffer (PH =6.8) was added to neutralize NaOH. In liquid medium, a mycobacterial culture supplement was mixed with a collection of antimicrobials called PANTA drugs, which are used to support the growth of mycobacteria and inhibit other microorganisms. The tubes for the solid culture were incubated at 37°C in a slant position to ensure an even distribution of inoculums for 1 week and thereafter in a straight position for another 7 weeks. The tubes were checked once every week for mycobacterial growth. The MGIT tubes were inoculated with an automated MGIT 960 culture system in which the machine detects the growth of oxygen consuming bacteria every hour. The machine indicates the presence of growth through UV-fluorescence. The MGIT culture was followed for 42 days, when thereafter, the machine automatically considers it as negative. The machine shows only the presence of growth but not the bacterial isolate. Print out results for both culture positive and negative results were accessed through the scanned bar code of the tubes.
After the MGIT machine showed light signaling, liquid cultures were subcultured on Brain Heart Infusion agar plates and incubated for 48 hours at 37°C to identify potential contaminants. All of the positive tubes were confirmed by ZN staining methods and the MPT64 protein-specific detection immune chromatographic test (SD Bioline Kit, Standard Diagnostics, Inc., Korea). Those isolates that were AFB positive, BHI growth negative, and SD bioline positive were confirmed as MTBC. The isolates that were AFB positive, BHI growth negative, and SD bioline negative were confirmed as non-tuberculosis mycobacteria (NTM).11
Phenotypic Drug Susceptibility Test
Isolates were tested for five first-line drugs: STM, INH, RIF, ETH, and PZA, using the MGIT 960 system. The phenotypic DST was performed using the proportional method recommended by the WHO. The concentration of drugs was 1.0 µg/mL, 0.1 µg/mL, 1.0 µg/mL, 5 µg/mL, and 100 µg/mL for STM, INH, RIF, EMB, and PZA, respectively. The tubes were incubated for a maximum of 13 days for STM, INH, RIF, and EMB, and for 21 days for PZA. Second-line DST was performed for MDR-TB isolates using the MGIT 960 system. For second-line DST, all liquid MTBC cultures that tested positive for MGIT within 1 to 5 days were used. A working solution of each drug was used at the concentration level of 2.0 μg/mL, 2.5 μg/mL, 0.25 μg/mL, 1.0 μg/mL, 1.0 μg/mL, 0.06 μg/mL, and 1.0 μg/mL for ofloxacin (OFX), levofloxacin (LFX), moxifloxacin (MOX), linezolid (LZD), bedaquiline (BDQ), delamanid (DLM), and clofazimine (CFZ), respectively, based on the WHO recommendations.12
Genotypic Drug Susceptibility Test
Genotype MTBDRplus and MTBDRsl assays (line probe assay) were performed directly on the MTB isolates according to the manufacturer’s instructions (Hain Lifescience, Nehren, Germany).13 The testing process included DNA extraction, master mix preparation, PCR, and reverse hybridization, which were done as part of the process, which was all performed in separate rooms. The person performing the tests was blinded to the phenotypic DST. It was considered a valid result if all expected control bands appeared valid; otherwise, the result was considered invalid. The absence of at least one of the wild-type bands or the presence of bands indicating a mutation in each drug resistance-related gene implied that the sample was resistant to a specific antibiotic. When all wild-type probes of a gene stained positive and there were no detectable mutations within the region examined, the sample was considered susceptible to the respective antibiotic.
Discard of Biological Sample
To minimize the risk of all infectious materials (processed sample), the samples were autoclaved before disposal. Autoclaves were available in the laboratory where TB culture is performed, Figure 1.
 |
Figure 1 Work flow on M. tuberculosis isolation, phenotypic and genotypic drug resistance pattern among EPTB patients in Addis Ababa, Ethiopia, 2022. |
Data Analysis
Data were analyzed using SPSS version 23 software (IBM Corporation, NY, Chicago). Descriptive statistics were used to show quantitative data. Binary and multinomial logistic regression analyses were conducted to determine the association of independent variables with the dependent variable. The significance level of P≤0.05 was considered statistically significant.
Ethical Consideration and Consent
The protocol for this study was approved by the IRB of SPHMMC and Addis Ababa Public Health Research and Emergency Management Directorate. The number and date of ethical approval letter was pm23/427 and 04/03/2022, respectively. The study was done as per the Helsinki declaration. Written consent was obtained from each study participant. Participants with drug-resistant isolate had their results forwarded to clinicians for a better treatment option.
Results
Source of Isolates and Data Collection
Using a pretested, structured questionnaire, both the socio-demographic and clinical data were collected by trained data collectors. Isolates from study participants were subjected to genotypic and phenotypic drug resistance pattern tests. The total number of MTBC isolates identified and subjected to drug susceptibility testing was 44. The isolates were obtained from 308 study participants.
Socio-Demographic Characteristics of Participants
Of 308 study participants included in the study, more than half (51.9%) were females. The mean age of the participants was 35.74 years, with a standard deviation of ±17.73 years. The majorities, 167 (54.2%) were married, 89 (28.9%) were illiterate, and 207 (67.2%) came from urban settings. Of 308 participants, 44 (14.2%) were with culture-positive cases, with the majority, 28 (63.6%) male proportion. The majority of positive cases, 23 (52.3%)-were in the age group of 21–40 years. The mean age of the participants was 34.20 years, with a standard deviation of ±15.33 years. The majorities, 26 (59.1%) were married, 18 (40.9%) had primary level education, and 32 (72.7%) were from urban settings. The socio-demographic characteristics of confirmed EPTB patients are summarized in Table 1.
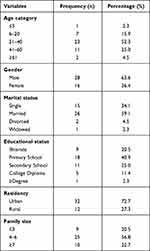 |
Table 1 Socio-Demographic Characteristics of the Participants (n=44) |
Clinical Characteristics of Participants
Of 44 culture-positive study participants, 34 (77.3%) were new presumptive patients, while 10 (22.7%) had a disease relapse. From the participants, 11 (25.0%) had known contact with TB cases. The majority, 31 (70.5%) positive cases, were TBLN, followed by pleura, which accounted for 6 (13.6%). Of the total culture-positive cases, 7 (15.9%) were HIV positive, and 7 (15.9%) had known COVID-19 disease history. The clinical characteristics of culture-positive patients are summarized in Table 2.
 |
Table 2 Clinical Characteristics of the Participants (n=44) |
Phenotypic Drug Susceptibility Test
After identification, 44 MTBC isolates were subjected to phenotypic DST with first-line anti-tubercular drugs. The extent of any drug resistance was 11/44 (25%). Isoniazid and PZA mono-resistance were the most common patterns, accounting for 6/44 (13.6%) and 5/44 (11.4%) of the isolates, respectively. Rifampicin resistance was found in two isolates (4.5%). Two of the isolates tested were resistant to ETH and STM (one isolate for each). MDR-TB was discovered in two of the RIF-resistant isolates, accounting for 4.5% of the isolates. MDR-TB isolates were subjected to second-line DST, but no XDR or pre-XDR isolates were discovered. Some isolates showed resistance to more than one drug. Dual-resistance patterns to INH and RIF (MDR-TB), INH and PZA, INH and STM, and PZA and EMB were seen in 2/44 (4.5%), 1/44 (2.3%), 1/44 (2.3%), and 1/44 (2.3%) of the isolates, respectively. Surprisingly, one isolate proved resistant to all first-line drugs. The majority of the resistant TB isolates (90.9%) came from new patients and TBL (63.6%). The phenotypic resistance pattern of the isolates is summarized in Table 3.
 |
Table 3 Phenotypic Drug Resistance Patterns of the Isolates for First-Line Drugs (n=44) |
Genotypic Drug Susceptibility Test
A genotype-based first-line drug sensitivity test was performed on 44 MTBC isolates. Of the 44 isolates, 5 (11.4%) were INH-resistant, while 2 (4.5%) were RIF-resistant. From the total cases, 3 (6.8%) were INH mono-resistant. The overall frequency of MDR-TB among the 44 isolates was 2 (4.5%). All RIF resistant isolates had rpoB mutations in codons 530–533, which showed the absence of the wild-type band (WT8) had an amino acid mutation at the S531L position 2 (4.5%). Mutations in the katG and inhA genes lead to INH resistance. In this study, all INH resistance mutations were in the katG gene. Of the 44 isolates, 5 (11.4%) were missing from the wild-type band (WT) at codon 315, 3 (6.8%) had an amino acid mutation at the S315T1 position, and one (2.3%) isolate had one at the S315T2 position. The MDR-TB isolates were tested for genotypic second-line drug susceptibility. However, no XDR or pre-XDR isolates were found. The frequency of gene mutations associated with rifampicin and isoniazid resistance among presumptive extrapulmonary patients is summarized in Table 4.
 |
Table 4 Frequency of Gene Mutations Associated with Rifampicin and Isoniazid Resistance (n=44) |
Associated Factors for Drug Resistance
In this study, various parameters were evaluated for their association with infection with drug-resistant tuberculosis. Gender, residency, family size, alcohol consumption, case type, history of anti-TB treatment, aspiration type, HIV status, and the history of COVID-19 were tested for any statistically significant association. None of the variables was found to be associated with drug-resistant tuberculosis. Bivariate and multivariate logistic regression analyses are summarized in Table 5.
 |
Table 5 Bivariate and Multivariate Analysis of Participants (n=44) |
Discussion
Based on the phenotypic drug susceptibility testing, The burden of any drug resistance among the isolates was found to be 11 (25%). Isoniazid and PZA mono-resistance were the most common resistance patterns, accounting for 6 (13.6%) and 5 (11.4%) of the isolates, respectively. The finding of any drug resistance in this study was in agreement with the study in Indonesia (22%).14 The finding of any drug resistance is lower than the studies reported in Northern India (39.9%), Southwest China (30.8%), and Cambodia (34.7%).3,15,16 The lower drug resistance rate of our study could be due to the study participants, who were entirely HIV patients, differences in the availability of drug types, and differences in TB control programs. Our finding was higher than the reports in Ghana (15.5%), Qatar (6.7%), and Addis Ababa (19.2%).17–19
The magnitude of MDR-TB in our study (4.5%) was higher than that reported from Indonesia (0.8%) and Qatar (1.2%).14,18 The higher MDR-TB rates in this study than in Indonesia and Qatar could be due to geographical and socio-economic differences. Our MDR-TB magnitude was lower than the findings from North India (13.4%), Southwest China (7%), Cambodia (8.1%), and surveillance from reference laboratories in Ethiopia (11.6%) and Addis Ababa (9%).3,15,16,19,20 The lower rate of MDR-TB than that of Southwest China and Cambodia could be due to the fact that the participants in the mentioned studies were entirely HIV positive patients. The study conducted at reference laboratories was done on stored isolates, which may be the reason for the variation. There were relatively fewer isolates in this study than in the previous study done in Addis Ababa.
Among the 44 isolates tested using the Geno Type MTBDRplus assay, 5 (11.4%) were INH resistant and 2 (4.5%) were RIF-resistant. From the total isolates, 3 (6.8%) were INH mono-resistant. All RIF resistant isolates had rpoB mutations at codon 530–533, which showed the absence of the wild-type band (WT8) with the appearance of MUT and an amino acid mutation at the S531L position 2 (4.5%). Mutations in the katG and inhA genes lead to INH resistance. In this study, all INH resistance mutation patterns were in katG. Of the 44 isolates, 5 (11.4%) were missing from the wild-type band (WT) at codon 315, 3 (6.8%) had an amino acid mutation at the S315T1 position, and one (2.3%) isolate had one at the S315T2 position. The MDR-TB isolates were tested for genotypic second-line drug susceptibility. However, no XDR or pre-XDR isolates were found. The higher proportion of INH resistance in the genotypic DST and the presence of mutations in the katG gene with a mutation at codon 315 and rpoB gene mutation at S531L were supported by previous studies conducted in Addis Ababa.19,21,22 In the absence of horizontally acquired resistance determinants, chromosomal changes provide resistance to drugs active against M. tuberculosis. These chromosomal alterations may impart drug resistance by drug target change or overexpression, as well as by preventing prodrug activation.23
Our study has some limitations. This study only included participants from Addis Ababa, and therefore may not accurately reflect drug resistance patterns in EPTB patients throughout the country. The molecular characterization of Mycobacterial tuberculosis strains was not performed using advanced molecular diagnostic tools like whole genome sequencing. However, our findings provide important evidence on drug resistance patterns and mutation characteristic of EPTB.
Conclusion
This study has shown a high proportion of INH and PZA resistance. The overall prevalence of MDR-TB was also high. A higher mutation rate was detected at the katG and rpoB genes, which confer resistance to INH and RIF drugs, respectively. For early case detection and treatment, expanding diagnostic capacity for first-line DST is a vital step to limit further spread of drug resistant TB strains in the study area.
Abbreviations
AOR, Adjusted Odds Ratio; AMR, Antimicrobial Resistance; BDQ, Bedaquiline; COVID-19, Corona Virus Disease 19; CFZ, Clofazimine; COR, Crude Odds Ratio; DLM, Delamanid; DM, Diabetes Mellitus; DNA, Deoxyribonucleic Acid; DST, Drug Susceptibility Test; EMB, Ethambutol; EPHI, Ethiopian Public Health Institute; EPTB, Extra pulmonary Tuberculosis; ETH, Ethionamide; FNA, Fine Needle Aspiration; HIV, Human Immunodeficiency Virus; INH, Isoniazid; IRB, Institutional Review Board; LFX, Levofloxacin; LPA, Line Probe Assay; LZD, Linezolid; MFX, Moxifloxacin; M. tuberculosis, Mycobacterium tuberculosis; MGIT, Mycobacterium Growth Indicator Tube; MTBC, Mycobacterium tuberculosis Complex; MDR, Multi-drug Resistance; MUT, Mutant; NTRL, National Tuberculosis Reference Laboratory; OFX, Ofloxacin; PCR, Polymerase Chain Reaction; PZA, Pyrazinamide; RIF, Rifampicin; RR, Rifampicin Resistant; SPHMMC, St. Paul’s Hospital Millennium Medical College; SPSS, Statistical Package for Social Science; STM, Streptomycin; TBLN, Tuberculosis Lymphadenitis; WT, Wild Type; WHO, World Health Organization; XDR, Extensively Drug Resistant.
Data Sharing Statement
All relevant data are within the manuscript.
Acknowledgment
The authors would like to acknowledge St. Paul’s Hospital Millennium Medical College, and the Ethiopian Public Health Institute for providing materials and facilities during this study. The authors also extend their gratitude to the study participants.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229–241. doi:10.1177/2042098614554919
2. Iacobino A, Fattorini L, Giannoni F. Drug-Resistant Tuberculosis 2020: where We Stand. Appl Sci. 2020;10:1–17.
3. Wang DM, Feng LQ, Zhu M, Xu YH, Liao Y. Clinical characteristics, common sites and drug resistance profile in culture-confirmed extrapulmonary TB/HIV co-infection patients, Southwest China. J Glob Antimicrob Resist. 2022;28:1–7. doi:10.1016/j.jgar.2021.10.028
4. Otu J, Gehre F, Zingue D, et al. Multidrug-Resistant Tuberculosis (Mdr-Tb): an Emerging Problem in West Africa. BMJ Glob Heal. 2017;2:A32.3–A33. doi:10.1136/bmjgh-2016-000260.85
5. Prasad R, Gupta N, Singh M. Multidrug resistant tuberculosis: trends and control. Indian J Chest Dis Allied Sci. 2014;56:237–246.
6. Lange CH, Chesov DU, Heyckendorf JAN, Leung CHIC. Invited Review Series: Tuberculosis Updates 2018. Drug-Resistant Tuberculosis. 2018:656. doi:10.1111/resp.13304
7. Singh PK. Epidemiological perspective of drug resistant extrapulmonary tuberculosis. World J Clin Infect Dis. 2015;5:77. doi:10.5495/wjcid.v5.i4.77
8. Gambhir S, Ravina M, Rangan K, Dixit M, Barai S. Imaging in extrapulmonary tuberculosis. Int J Infect Dis. 2017;56:237–247. doi:10.1016/j.ijid.2016.11.003
9. Schön T, Miotto P, Köser CU, Viveiros M, Böttger E, Cambau E. Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin Microbiol Infect. 2017;23:154–160. doi:10.1016/j.cmi.2016.10.022
10. Dusthackeer A, Sekar G, Chidambaram S, Kumar V, Mehta P, Swaminathan S. Drug resistance among extrapulmonary TB patients: six years experience from a supranational reference laboratory. Indian J Med Res. 2015;142:568–574. doi:10.4103/0971-5916.171284
11. Tortoli E, Cichero P, Piersimoni C, Simonetti MT, Gesu G, Nista D. Mycobacteriology Laboratory Manual. J Clin Microbiol. 2014;37:3578–3582. doi:10.1128/jcm.37.11.3578-3582.1999
12. Yousif N, Cole J, Rothwell JC, et al. Technical Manual for Drug Susceptibility Testing of Medicines Used in the Treatment of Tuberculosis. Vol. 9. World Health Organization; 2018.
13. Armstrong D. Hain genotype line probe assay: overview and training. Int J Med. 2018;17.
14. Cucunawangsih WV, Widysanto A, Lugito NPH. Mycobacterium tuberculosis resistance pattern against first-line drugs in patients from urban area. Int J Mycobacteriol. 2015;4:302–305. doi:10.1016/j.ijmyco.2015.08.002
15. Walls G, Bulifon S, Breysse S, et al. Drug-resistant tuberculosis in HIV-infected patients in a national referral hospital, Phnom Penh, Cambodia. Glob Health Action. 2015;8:2–10. doi:10.3402/gha.v8.25964
16. Maurya AK, Kant S, Kushwaha RAS, Dhole T, Nag VL. Trends of anti-tuberculosis drug resistance pattern in new cases and previously treated cases of extrapulmonary tuberculosis cases in referral hospitals in northern India. J Postgrad Med. 2012;58:185–189. doi:10.4103/0022-3859.101379
17. Addo SO, Mensah GI, Mosi L, Abrahams AOD, Addo KK. Genetic diversity and drug resistance profiles of Mycobacterium tuberculosis complex isolates from patients with extrapulmonary tuberculosis in Ghana and their associated host immune responses. IJID Reg. 2022;4:75–84. doi:10.1016/j.ijregi.2022.06.005
18. Ali M, Howady F, Munir W, et al. Drug-resistant tuberculosis: an experience from Qatar. Libyan J Med. 2020:15. doi:10.1080/19932820.2020.1744351
19. Getu D, Kebede A, Habteyes Hailu Tol BY. Molecular characterization and drug resistance patterns of Mycobacterium tuberculosis complex in extrapulmonary tuberculosis patients in Addis Ababa, Ethiopia. PLoS One. 2020;29:1–13. doi:10.1371/journal.pone.0243493
20. Diriba G, Kebede A, Tola HH, et al. Surveillance of drug resistance tuberculosis based on reference laboratory data in Ethiopia. Infect Dis Poverty. 2019;8:4–9. doi:10.1186/s40249-019-0554-4
21. Zewdie O, Mihret A, Abebe T, et al. Genotyping and molecular detection of multidrug-resistant Mycobacterium tuberculosis among tuberculosis lymphadenitis cases in Addis Ababa. New Microbes New Infect. 2018;21:36–41. doi:10.1016/j.nmni.2017.10.009
22. Diriba G, Kebede A, Tola HH, et al. Utility of line probe assay in detecting drug resistance and the associated mutations in patients with extrapulmonary tuberculosis in Addis Ababa, Ethiopia. SAGE Open Med. 2022;10:205031212210982. doi:10.1177/20503121221098241
23. Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41:354–373. doi:10.1093/femsre/fux011
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
