Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Pharmacogenomics and Personalized Medicine in Type 2 Diabetes Mellitus: Potential Implications for Clinical Practice
Authors Venkatachalapathy P , Padhilahouse S , Sellappan M , Subramanian T , Kurian SJ, Miraj SS, Rao M, Raut AA, Kanwar RK, Singh J, Khadanga S, Mondithoka S, Munisamy M
Received 24 July 2021
Accepted for publication 4 October 2021
Published 13 November 2021 Volume 2021:14 Pages 1441—1455
DOI https://doi.org/10.2147/PGPM.S329787
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Martin H Bluth
Poongothai Venkatachalapathy,1 Sruthi Padhilahouse,1 Mohan Sellappan,1 Tharunika Subramanian,2 Shilia Jacob Kurian,3 Sonal Sekhar Miraj,3 Mahadev Rao,3 Ashwin Ashok Raut,4 Rupinder Kaur Kanwar,4 Jitendra Singh,4 Sagar Khadanga,5 Sukumar Mondithoka,5 Murali Munisamy4
1Department of Pharmacy Practice, Karpagam College of Pharmacy, Coimbatore, Tamilnadu, India; 2Karpagam Faculty of Medical Sciences and Research, Coimbatore, India; 3Department of Pharmacy Practice, Manipal College of Pharmaceutical Sciences, Manipal Academy of Higher Education, Manipal, Karnataka, India; 4Translational Medicine Centre, All India Institute of Medical Sciences, Bhopal, Madhya Pradesh, India; 5Department of General Medicine, All India Institute of Medical Sciences, Bhopal, Madhya Pradesh, India
Correspondence: Murali Munisamy
Translational Medicine Centre, All India Institute of Medical Sciences, Bhopal, 462020, Madhya Pradesh, India
, Tel +91-9600-883-284
Email [email protected]
Abstract: Type 2 diabetes mellitus (T2DM) is the most common form of diabetes, and is rising in incidence with widespread prevalence. Multiple gene variants are associated with glucose homeostasis, complex T2DM pathogenesis, and its complications. Exploring more effective therapeutic strategies for patients with diabetes is crucial. Pharmacogenomics has made precision medicine possible by allowing for individualized drug therapy based on a patient’s genetic and genomic information. T2DM is treated with various classes of oral hypoglycemic agents, such as biguanides, sulfonylureas, thiazolidinediones, meglitinides, DPP4 inhibitors, SGLT2 inhibitors, α-glucosidase inhibitors, and GLP1 analogues, which exhibit various pharmacogenetic variants. Although genomic interventions in monogenic diabetes have been implemented in clinical practice, they are still in the early stages for complex polygenic disorders, such as T2DM. Precision DM medicine has the potential to be effective in personalized therapy for those suffering from various forms of DM, such as T2DM. With recent developments in genetic techniques, the application of candidate-gene studies, large-scale genotyping investigations, genome-wide association studies, and “multiomics” studies has begun to produce results that may lead to changes in clinical practice. Enhanced knowledge of the genetic architecture of T2DM presents a bigger translational potential. This review summarizes the genetics and pathophysiology of T2DM, candidate-gene approaches, genome-wide association studies, personalized medicine, clinical relevance of pharmacogenetic variants associated with oral hypoglycemic agents, and paths toward personalized diabetology.
Keywords: pharmacogenomics, personalized medicine, type 2 diabetes, antidiabetic drugs
Introduction
Type 2 diabetes mellitus (T2DM), a complex polygenic disorder, is a major burden worldwide.1 Genome-wide association studies (GWASs) have detected several gene variants associated with diabetes in different Indian subethnic populations. Population-specific riskalleles have been seen to increase diabetes prevalence in South Asians.2 The worldwide prevalence of diabetes has been predicted to double from 171 million cases in 2000 to 366 million in 2030, and then to 642 million by 2040, with approximately 79.4 million by 2030 in India.3 According to Wild et al, the “top” three countries with the most T2DM cases are India, China, and the US, with estimates of 79.4 million, 42.3 million, and 30.3 million by 2030, respectively.4 Although diabetes is a global health concern, its burden is more evident in developing countries like India. Economically, the global encumbrance of diabetes is huge, with 75% prevalence in low- and middle-income countries (LMICs). India is considered the diabetes capital of the world, with a large number of diabetic subjects and individuals remaining undiagnosed, accounting for >50% of people.3 The use of molecular testing to customize treatment widely is not yet possible. Furthermore, diabetes treatment based on a homogeneous therapeutic algorithm frequently leads to therapeutic failure with various diabetic complications.3 With the advancement of high-throughput sequencing technologies, combined “omics” data, such as genomics, transcriptomics, proteomics, metabolomics, can be accumulated and used in global profiling of health and diseases.4 Combined analysis of big data and routinely gathered clinical and laboratory data can be used in personalized therapeutic approaches.5 Personalized medicine is the most promising strategy in treating a complicated polygenic illness like T2DM, because of variability in phenotypes across population groups and the need to determine the appropriate medication for each individual.6 This new paradigm is based on the patient’s genetic and metabolic structure to customize diabetes diagnostics, prevention, prognostics, and treatment. Comprehending the widespread prevalence of diabetes, personalized diabetes management is considered imperative. As such, this demands the development and implementation of a framework for personalized diabetes care. The road to personalized medicine is interesting, yet challenging. This review focuses on the current opportunities and challenges for implementation of personalized medicine in the clinical practice of T2DM management — “personalized diabetology.”
Diabetes Pathogenesis and Gene Variants
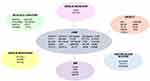
GWASs have identified several gene loci involved in the various pathophysiological pathways of diabetes, explaining its complex polygenic nature.7–9 Various gene loci are involved in insulin secretion, insulin resistance, obesity-associated diabetes, fasting glucose, β-cell count, and function. These genomic data can help in early disease prevention and selection of tailored diabetic therapy to achieve optimal glycemic control, thereby preventing or delaying the development of diabetic complications. In Figure 1, the pathogenic effects of certain T2DM-related genes in Indian populations are summarized, based on GWASs on T2DM pathogenesis in Indian subjects.10–32
Genetics of Type 2 Diabetes
Candidate-Gene Studies
The candidate-gene approach focuses on a population of distinct individuals, rather than related family members. These studies are theory-motivated,1 analyzing gene variants within functional candidate genes based on data generated by linkage studies regarding genetic association. Though novel genes cannot be identified,1 these studies signify as the most influential method. Intensive sequencing of genes thought to be involved in T2DM pathogenesis like glucose metabolism, insulin secretion, and insulin resistance is done in candidate genetic analysis.33 Along with the assistance of data from the Human Genome Project, which includes a public database of single-nucleotide polymorphisms (SNPs), candidate genetic variants are detected.
PPARG
An initial candidate gene positively associated with T2DM was PPARG of the nuclear hormone–receptor family, regulating transcription.1 As a molecular target for the antidiabetic-drug class thiazolidinediones, this makes it a promising candidate gene.34 Substitution of proline for alanine at position 12 in this protein, ie, polymorphism Pro12Ala (rs1801282), in PPARG2 on extra exon B has been observed to yield a 20% higher risk of diabetes.33 The genetic variant in this gene has been found to have high correlation with elevated transcriptional function, and an elevated function of defense against T2DM.35
KCNJ11
KCNJ11 is an inwardly rectifying potassium channel (subfamily J, member 11) encoding Kir6.2. It is an ATP-sensitive channel, coding for four subunits.34 It acts as a significant gene in regulation of insulin secretion by β cells,33 where polymorphisms lead to elevated K-ATP channel function, causing β-cell dysfunction.36 In 1998, a missense polymorphism in KCNJ11 E23K was initially identified to be related to T2DM and confirmed by various studies, including GWASs.37 KCNJ11 is associated with neonatal diabetes as well, and its rare potential polymorphism can even lead to a permanent form of neonatal diabetes.38
IRS1 and IRS2
Insulin Receptor Substrate 1 and Insulin Receptor Substrate 2 (IRS1 and IRS2) play a crucial role in the insulin-signaling cascade,39 and polymorphisms in these genes have been found to be linked with reduced insulin sensitivity.40
WFS1
The missense mutation rs734312 is found in exon 8 of Wolfarin ER Transmembrane Glycoprotein (WFS1). Also, elevated oral glucose-tolerance test–derived insulin-secretion levels are related to variant rs10010131. These two polymorphisms in have been found to have substantial defensive action against T2DM.41 SNPs in WFS1 have strong associations with T2DM.
HNF1A, HNF1B, and HNF4A
HNF1A, HNF1B, and HNF4A are significantly associated with monogenetic diabetes in the young, also called maturity-onset diabetes of the young.33 The 127L, A98V, and S487N variants in HNF1A mutation have decreased transcription function in genes engaged in GLUT2 mechanisms. Polymorphisms of HNF1A like AG8V and S487N are highly developed in late-onset autosomal-dominant DM, which is clinically similar to T2DM.42
ENPP1
ENPP1 is associated with T2DM.43 The missense variant rs1044498 of the ENPP1 K121Q polymorphism is associated with T2DM and the development of insulin resistance, which was also supported by various other studies in distinct populations.44 A meta-analysis on 11,855 Chinese subjects established that the Q allele of K121Q gene may act as a predisposing factor of T2DM, augmenting T2DM susceptibility.45 However, no association has been replicated in other studies on different populations such as a one involving north Indian subjects, which reported no associations among ENPP1 K121 polymorphisms, T2DM, and related quantitative metabolic traits.46
Genome-Wide Association Studies
The Human Genome Project, completed in 2003, mapped the entire human genome.47 This has led to subsequent developments in genomic research. The international haplotype map (HapMap) project primarily sequenced 3.9 million SNPs in 270 DNA samples from four distinct ethnic populations, followed by detection of millions of SNPs, which got stored on a public database.48 Another international research effort, the 1000 Genome Project, has also detected SNPs throughout the human genome and added data, and is used widely by the research community. Utilization of these data sources and enhancement of advanced high-throughput sequencing technology thus play an important role in studying various T2DM-associated genes and in comprehending the disease at its genetic level. A French cohort study involving 661 T2DM cases and 614 controls that covered 3,92,935 SNP loci was the first GWAS to identify novel genetic variants like SLC30A8, HHEX, EXT2, and COC387761 as being associated with T2DM.49 GWASs have illustrated novel pathways, pointed toward fundamental biology, confirmed prior epidemiological observations, drawn attention to the role of β-cell dysfunction in T2DM, explained ~10% of disease heritability, tempered our expectations with regard to their use in clinical prediction, and provided possible targets for pharmacotherapy and pharmacogenetic clinical trials. GWASs have also been integrated with high-throughput metabolomic profiling to provide scientific insights into how genetic diversity influences metabolism and how metabolic differences in plasma might help identify important genes within chromosomal areas associated with T2DM.50
Personalized Medicine: A Paradigm Shift in Diabetes Treatment
Applying data generated from various clinical trials on the genetics of diabetes involving subjects who are usually young with few or no comorbid diseases to the general diabetic population remains a challenge. Even with data produced from individuals meeting selective inclusion criteria of glycemic control and development of complications, replicating this evidence-based medicine for diabetic patients of various heterogeneity may not always provide a similar outcome. Sometimes, it even leads to adverse outcomes. With diverse genetic variants studied in GWASs, linkage with different diabetic risks and pathogenesis mechanisms like insulin secretion and resistance, glucose homeostasis, and membrane transportation necessitates personalized medicine in diabetes management.51
In complex polygenic disorders like T2DM, early risk prediction and prevention are essential. Various randomized controlled trials have established that the risk of developing diabetes can be reduced by half if predicted early. Personalized medicine can play a potential role, enabling clinicians to provide tailored therapy.52 In addition to clinical markers like phenotypic characteristics and markers of metabolism, endothelial dysfunction markers, data on well-established genetic variants associated with T2DM risk possess great significance in diabetic prevention. Genetic variants in TCF7L2, PPARG, KCNJ11, WFS1, SLC30A8, JAZF1, and HNF1B have been established as posing a high risk of developing T2DM.28 Therefore, the use of big data generated by GWASs and other “multiomics,” including proteomics, metabolomics, and transcriptomics, along with advanced high-throughput sequencing technologies, will provide a promising future in precision medicine for diabetes. Although, genetic testing regarding the monogenic form of diabetes is available as a tool in specialized diabetes clinics, the use of precision medicine for the polygenic form of diabetes has not yet evolved. Incorporating omics data with clinical phenotype data of a patient potentially aids in better risk prediction, prevention, and management of T2DM. A recent data-driven cluster analysis of six diabetes-related variables in newly diagnosed diabetes patients from the Swedish All New Diabetics in Scania cohort (n=8,980) has been replicated in three other cohorts: the Scania Diabetes Registry (n=1,466), All New Diabetics in Uppsala (n=844), and Diabetes Registry Vaasa (n=3,485). Five clusters of diabetic patients with distinct disease characteristics and a higher risk of diabetic complications were identified, each with a different genetic association from conventional T2DM. As a result, such subcategorization aided in a better understanding of diabetes stage and pathogenesis, allowing for targeted and early intervention. This new substratification might eventually help to tailor and target early treatment for patients who would benefit most, thereby representing a first step toward precision medicine in diabetes.53
Deep-learning algorithms, which can detect exceedingly complex patterns in huge data sets, have been shown to be effective in illness-prediction models and biological process prediction.54 These findings demonstrate that a multiomics technique provides additional information for T2DM prediction and treatment management. In the near future, deep-learning algorithms may be applied in multiomics studies on T2DM, as well as precision medicine. The development of systems biology methods for the integration of multiomics data is crucial for forecasting rising fasting plasma-glucose levels. SNPs in such genes as RPL7AP27, SNX30, SLC39A12, and BACE2 have been found to be highly associated with increased fasting plasma-glucose levels.55 This demonstrates that combining candidate SNPs with IgG glycomics can yield T2DM-biomarker potential. The strong predictive potential observed by integrating genomes and glycomic biomarkers suggests that such multiomic approaches could be used to provide predictive, preventive, and personalized T2DM medication.
Effect of Pharmacogenetics on Antidiabetic Medications
Pharmacogenomics means formulating a genetically tailored therapeutic plan to achieve the best optimal individual response. The individual’s genetic profile is considered to optimize pharmacokinetics and pharmacodynamics, in achieving the desired drug efficacy and response. In the recent years, several gene polymorphisms on the therapeutic response of various anti-diabetic drugs have been studied. However, issues like lack of knowledge on clinical relevance and implementation, lack of structured guidelines and ethical, social, technological, legislative, and economic issues remains a challenge. Therefore, giving importance to interindividual genetic variability in response to antidiabetic agents is the primary factor in achieving “personalized diabetology”.
Metformin
ATM
A meta-analysis of three cohort studies — Hoorn Diabetes Care System (DCS) cohort,56 CARDS cohort,57 and smaller Rotterdam Study cohort58 — concluded that the ATM, a member of the PI3K family and important for cell-cycle control and DNA repair, in which rs11212617 polymorphism was associated with metformin-treatment response.59 This polymorphism and rs628031 of SLC22A1 were found to have no association with metformin treatment in an Iranian T2DM population.60 In a Caucasian population, rs11212617 had a significant association with metformin response, with low plasma concentration of metformin indicating high cellular-level action.61 However, in a south Indian population, these SNPs were found to have no contribution to T2DM incidence.62
OCT1
The allele and genotypes of the SLC22A1 rs622342 polymorphism were associated with metformin effectiveness in south Indian patients with T2DM.63 The GoDART database study examined rs122083571 and rs72552763 in 2,216 participants and reported that patients with these polymorphisms on OCT1 inhibitors had more than fourfold the risk of acquiring intolerance to metformin (OR 4.13, 95% CI 2.09–8.16; P<0.001).64 The rs2297374 polymorphism (+43C>T) and metformin response showed no significant association in Indian populations,65 and 20% frequency of rs2282143 (1022C>T) was detected in Indian subjects. The influence of rs1867351 (156T>C) on metformin-action regulation has been examined in an Indian population, showing a frequency of 27% (2018).66 A study on a Mexican population recently identified CC-rs622342 (β=1.36, P<0.001), AA-rs628031 (β=0.98, P=0.032), and GG-rs594709 (β=1.21, P=0.016) in the SLC22A1 gene to be associated with reduced metformin effectiveness, with increased HbA1c levels.67 The variants R61C (rs12208357), G401S (rs34130495), G456R (rs34059508), and 420del (rs72552763) were associated with reduced metformin activity.68
OCT2
Genetic variants in the SCL22A2 gene encoding the OCT2 protein, such as T199I, T201M, and A270S, have been fouund to be related to decreased metformin function.69 However, no significant association between SLC22A2 SNPs (rs10755577, rs17588242, rs17589858, rs2928035, rs312024, rs312025, rs312026, rs3127573, rs533452, and rs662301) and metformin clearance has been found in healthy Caucasian males.70 A recent study also failed to replicate associations between any SNPs of SLC22A2 and glucose regulation. However, using multinomial logistic regression and adjusting for covariates like age and BMI, associations between glucose regulation and SNPs within SLC22A1, SLC22A2, and SLC22A3 were replicated.71
OCT3
The four SLC22A3 SNPs (rs12194182, rs2292334, rs2504927, and rs3123634) have been found to have no association with metformin action in Caucasians.71 The rs2292334 and rs12194182 SNPs are associated with lower risk of T2DM and lower mean HbA1c levels. In an Iranian study, metformin showed better glucose regulation and lipid management, irrespective of OCT3-564G>A variant.72 The genetic variants in PRPF31, CPA6, and STAT3 are associated with novel glucose-lowering mechanisms for metformin.73 A significant association was observed in a recent study in 2019 on T2DM patients between TCF7L2 rs7903146 and metformin response.74 Carriers of the G allele of the intronic SNP rs3889348 exhibit significantly lower expression of SLC29A4, which encodes PMAT. Since it aids in metformin absorption, metformin therapy increases the risk of gastrointestinal intolerance.75
Sulfonylureas
Sulfonylureas are metabolized in the liver primarily by the polymorphic cytochrome P450 isoenzyme 2C9, encoded by CYP2C9. In a large GoDARTS64 retrospective study of 1,073 subjects, carriers of loss-of-function CYP2C9*2 or CYP2C9*3 alleles had 3.4-fold the higher probability of attaining glycemic control of carriers of the wild-type alleles. Two polymorphisms — CYP2C9*2 (I359L) and CYP2C9*3 (R114C) — were associated with elevated serum-sulfonylurea levels.76
Sulfonylureas are insulin secretagogues that bind the SUR1 subunit (encoded by ABCC8), play a major role in insulin secretion, and are potential candidate for T2DM. The 3c → t polymorphism and the Thr759Thr (ACC → ACT) silent polymorphism were initially associated with T2DM in Caucasians.77 A genotyping study assessing this polymorphism failed to replicate this in a south Indian population of 637 diabetes patients.78 The KCJN11 E23K variant is associated with T2DM and sulfonylurea efficacy in Caucasians.79 In Caucasian T2DM patients, rs7903146 and rs1801278 polymorphisms of the TCF7L2 and IRS1 genes are associated with poor sulfonylurea response.80 In an Indian study involving a Gujarat population of T2DM patients, genetic variation at rs12255372 was associated with the sulfonylurea effectiveness.81 Several genetic variants of TCF7L2 are related to T2DM in diverse ethnicities, among which rs7903146 (intron 4) has the strongest association with T2DM, while rs12255372 and rs7903146 are related to poor therapeutic outcomes.82,83 MIR4532 rs60452575 influenced KCNJ11 expression and sulfonylurea effectiveness in a Chinese population.84
DPP4 Inhibitors and GLP1 Analogues
DPP4 inactivates the incretins GLP1 and gastric inhibitory polypeptide (GIP). DPP4 inhibitors extend the half-life of these incretins, and this is correlated with augmented insulin release and reduced glucagon release.85 GLP1-receptor agonists and DPP4 inhibitors control blood glucose by targeting the body’s incretin system. GLP1 agonists act as “incretin mimetics” and DPP4 inhibitors prevent the breakdown of endogenous incretin. DPP4 inhibitors and GLP1-receptor agonists are recommended as second-line glucose-lowering agents by the American Diabetes Association and the European Association for the Study of Diabetes in cases where patients require combination therapy for adequate glycemic control or when metformin or sulfonylureas are ineffective.86,87 The first DPP4-selective inhibitor was sitagliptin, which was followed by vildagliptin, saxagliptin, linagliptin, and most recently alogliptin. Exenatide, liraglutide, lixisenatide, dulaglutide, and albiglutide are the five GLP1-receptor agonists currently approved for the treatment of T2D.88
In a recent study on a Central European population of 206 T2DM patients, missense variant rs6923761 in the GLP1R gene was associated with lower glucose control in 6-month exposure to gliptins.89 In individuals with high body fat, DPP4 rs6741949 in intron 2 position showed negative correlations with insulin secretion (P=0.0061), glucose tolerance (P=0.0208), and glucose-stimulated GLP1 levels (P=0.0229).90 The rs2285676 variant in the KCNJ11 gene is a predictor of the therapeutic effect of DPP4 inhibitors.91 In a study on 137 Caucasian diabetics, the KCNQ1 rs163184 T>G variant was related to glucose regulation of DPP4 inhibitors.92 Variants of CDKAL1 (rs7754840 and rs7756992) in Japanese are linked with glycemic control activity of DPP4 inhibitors.93 In the Taiwanese, rs57803087 in PRKD1 is highly associated with DPP4-inhibitor function.94 GLP1-analogue drugs are incretin mimetic agents. The SNP rs7202877 has been found to control the expression of CTRB1 and CTRB2 for chymotrypsin, a significant regulator of the incretin mechanism in non-T2DM patients.95 In a recent study, T2DM patients with minor A allele of GLP1R (rs6923761), who had received exenatide or liraglutide showed a more significant delay in gastric emptying T½ to baseline.96 Although TCF7L2 (rs7903146) and WFS1 (rs10010131) and KCNQ1 (rs151290, rs2237892, and rs2237895) were initially shown to be related to GLP1 response, another study of the effect of these variants on GLP1 concentrations showed no association in healthy individuals. Also, GLP1R polymorphisms showed no statistical association with GLP1-analogue responses in T2DM patients with poor glycemic control.97
Sodium–Glucose Cotransporter 2 Inhibitors
SGLT2 is encoded by the SLC5A2 gene, located on human chromosome 16p11.2. From genotyping of five SNPs in SLC5A2 gene locus in 603 T2DM subjects, no association between SLC5A2 variants and empagliflozin response was detected.98 On the other hand, the rs9934336 G allele has been found to be associated with increased 30-minute plasma glucose, 120-minute insulin concentrations, and AUC120 glucose on oral glucose-tolerance test in 907 nondiabetic Sorbs (P<0.05).99 In addition, the UGT1A9*3 and UGT2B4*2 polymorphisms have been demonstrated to increase plasma concentration of the SGLT2 inhibitor canagliflozin in carriers of wild-type alleles.100 Kan et al investigated the effect of alogliptin on liver function and glucose regulation in T2DM patients with nonalcoholic fatty-liver disease and PNPLA3 rs738409 C>G genotypes. Those with the G allele showed a positive relationship between improved HbA1c levels and alterations in liver-transaminase levels.101
α-Glucosidase Inhibitors
The STOP-NIDDM trial,102 with 770 study subjects, studied the acarbose response and its association with genetic variants of PPARA, HNF4A, LIPC, PPARG2, and PPARGC1A were studied. Findings were not replicated in other populations with preexisting T2DM. The Pro12Pro genotype of PPARG2 gene and the 482Ser allele of PPARGC1A has been established to be associated with the transformation of impaired glucose tolerance in T2DM. Acarbose averts the progression of diabetes, irrespective of PPARG2 genotype.103
Meglitinide
SLCO1B1, CYP2C8, CYP3A4, TCF7L2, SLC30A8, IGF2BP2, KCNJ11, KCNQ1, UCP2, NAMPT, MDR1, PAX4, and NEUROD1 were found to be associated with meglitinide response in the Chinese population.104 OATP1B1, which SLCO1B1 encodes, facilitates hepatic transport of the drug. Genetic polymorphisms in CYP2C8 and CYP2C8*1/*3 genotypes are associated with reduced plasma concentrations of repaglinide.105 In a study on Chinese T2DM patients on repaglinide, the NAMPT −3186C⁄T polymorphism affected plasma levels of postprandial serum insulin and total cholesterol levels.106 The KCNQ1 rs2237892 T and rs2237895 C alleles respond to repaglinide positively.107 As KCNQ1 plays a vital role in controlling insulin resistance through the IRS2–PI3K–Akt signaling pathway, the genetic polymorphism in this gene has been found to affect repaglinide response in the same population.107 The frequency of the ABCC8 rs1801261 allele has been found to be higher in T2DM patients than control subjects (22.6% vs 11%, P<0.01), exerting effects on repaglinide response.108 The C/C homozygotes of the ABCC8 exon16– 3T/C variant have shown better repaglinide response in insulin sensitivity than the T/C and T/T genotypes of the KCNJ11 E23K variant.109
Thiazolidinediones
Thiazolidinediones are PPAR activators that decrease circulating free fatty acids, thereby enhancing sensitivity to insulin and reducing hyperglycemic episodes.110 The rs296766 T allele of AQP2 and rs12904216 G of SLC12A1 have been found to be associated with edema in rosiglitazone users.111 PPARGC1A Thr394Thr and Gly482Ser polymorphisms are associated with rosiglitazone action in Chinese patients with T2DM.112 The P12A variant in PPARG is associated with lowered rosiglitazone effectiveness.113 Another Asian study with 250 patients demonstrated that carriers of the minor allele of variant rs1801282 in PPARG had higher odds of being responders to pioglitazone than carriers of wild-type alleles.114 Additionally, carriers of the A allele of rs6467136 in PAX4 showed improved response to rosiglitazone.115 The major metabolizer of thiazolidinedione is CYP2C8, in which the *3 variant116 has reduced response to insulin, with lower plasma concentration of rosiglitazone.117 The transporter OATP1B1, encoded by SLCO1B1, facilitates hepatic uptake of thiazolidinediones, which are metabolized by the enzyme CYP2C8 (encoded by CYP2C8), are associated with two variants — Val174Ala and rs4149056 — in the Scottish population.116 Genetic variants associated with therapeutic responses to antidiabetic medications are summarized in Table 1.
 |
Table 1 Genetic Variants that Influence Antidiabetic-Medication Response |
Current Perspectives and Future Prospects of Personalized Medicine in Type 2 Diabetes
The Precision Medicine in Diabetes Initiative was launched in 2018 by the American Diabetes Association in collaboration with the European Association for the Study of Diabetes and the US National Institute of Diabetes and Digestive and Kidney Diseases.118 Although the application of precision medicine in monogenic diabetes was successful, it is challenging to implement in T2DM, a complex multifactorial polygenic disease.
Over the years, more than 100 T2DM-susceptibility loci have been detected. However, the understanding of functions of these detected genetic variants in diabetic pathogenesis remains challenging. As the effect of causal variants in T2DM is small, it becomes hard to establish their association. This issue can be reduced using biobanks, which help in the accessibility of well-organized, multiuser, large-cohort databases covering clinical, laboratory, and molecular information from large patient samples. The DNA Technology Regulation Bill 2019 in India provides for the establishment of DNA data banks at national and regional levels. There are now 336 million people with diabetes living in LMICs,3 accounting for four in five people worldwide with diabetes. India, an LMIC that is a major epicenter of diabetes, is a diverse country with nearly 4,000 population groups and characterized by unique genetic variations within the subpopulations. GWASs involving a larger population of different ethnicities may lead to identification of more genetic loci associated with T2DM. They also may aid in the interpretation of the function and role of predetected genetic variants. This can be achieved as the cost of sequencing technologies reduces over time.
Establishing a set of biomarkers that would accurately associate with various stages of diabetes and complications is crucial. As molecular sequencing studies keep generating pharmacogenetic markers, clinical trials involving interventional therapies that target these should be conducted to ensure the reliability of the established data. One of the best examples of how precision medicine can be successfully exploited is sulfonylureas targeting the KCNJ11 genetic variation. Metformin has been observed to enhance the antitumor activity of MEK inhibitors in human LKB1 wild-type non–small cell lung cancer (NSCLC) cell lines, regardless of KRAS-mutation status, by downregulating GLI1 and decreasing NF-kB (p65)-mediated transcription of MMP2 and MMP9.119 The METAL trial was designed to determine the maximum tolerated dose and evaluate the safety and activity of metformin coupled with erlotinib in second-line treatment of patients with stage IV NSCLC whose tumors expressed the wild-type EGFR gene.120 A recent multicenter clinical trial on diabetic kidney disease called Nephropathy in Diabetes Type 2 compared standard of care (n=188) with multifactorial intensive therapy (n=207) in which comprehensive therapy for the main risk factors was far more effective than standard of care in preventing major fatal/nonfatal cardiovascular events in diabetic kidney-disease patients, and its use at an early stage offered prolonged protection. As a result, such an integrated and multifactorial approach may result in better diabetic outcomes.121 Sharing those trial results is crucial in providing new insights. Databases have been developed in recent years through sharing of data, such as the Human Gene Mutation Database122 and ClinVar.123
The ancillary effects of antiglycemic drugs can also be tailored and directed toward beneficial results. In major randomized clinical trials and real-world observational studies, SGLT2 inhibitors have shown positive pleiotropic effects on body weight, systolic blood pressure, and eGFR levels, as well as improved cardiovascular outcomes. These pleiotropic effects are advantageous for the prevention or decrease of macro- and microvascular problems, and may be especially beneficial in patients with diabetes or at risk of diabetes complications, such as CVD, HF, and CKD. This enables physicians to choose appropriate glycemic therapy based on cardiovascular and renal comorbidities.124,125
Electronic health-care records across health-care systems are crucial in implementing of precision medicine for diabetes, as they are easy to access and share among various systems across a wide region. Collaborations among various research societies, health-care organizations, funding organization, suppliers, and governing agencies to implement precision medicine in diabetes diagnostics, prevention, monitoring, prognostics, and treatment are crucial. It is essential to form an active network of stakeholders with patient representatives and public organizations to raise agendas and funds.
Although diabetes precision medicine involving sequencing technologies is more expensive than conventional treatment, precision medicine in monogenic diabetes has been established to be cost-effective. As diabetic complications are the primary factor in treatment expenses, early diagnosis, prevention, and intervention based on genetic variants through precision medicine may be motivation for acceptance. A critical evaluation of the cost versus benefit of sequencing technologies, genomics, and biomarkers is necessary to advocate its use in clinical practices in certain populations. The use of technology in diabetes, such as wearable glucose-monitoring sensors with minimal invasion and uninterrupted glucose measuring, is highly encouraged and practiced in various health-care systems, the best example of extensive personalized medicine in diabetes.126
Algorithms and guidelines on personalized diabetes therapy based on genotype should be developed based on the clinical evidence generated, aiding in implementing such evidence at the clinical level. The exploitation of artificial intelligence in clinical decision-making for an optimal therapeutic regimen for many patients will be the revolutionizing approach in personalized medicine of diabetes. Educational programs are required to train and educate clinicians, geneticists, and other health-care professionals in implementing personalized medicine for diabetes at the patient level and handling potential accidental findings, such as unexpected germ-line mutations. Adequate training of the genomic workforce can be achieved by procuring suitable funds for providing genomic education. The participation of regulatory bodies in the initial phases of precision-medicine development in diabetes is crucial for its effective execution and practice.
Individual genetic variation identification and knowledge of its role in the predisposition and pathogenesis of T2D would be a significant step in disease management, improving clinical conditions and preventing complications. In this review, we have identified the current state of genetic risk variants linked with T2DM and shown the importance pharmacogenomic studies have in associating actionable relationships between genetic and pharmacological treatments. Personalized medicine can lead to more effective drug therapy with better patient adherence in routine clinical practice. Precision DM medicine is already being used to treat monogenic forms, such as maturity-onset diabetes of the young, neonatal DM, and congenital hyperinsulinemic hypoglycemia. Precision DM medicine promises to be useful in customized therapy for those suffering from different types of diabetes, such as T2DM.127
T2DM is a polygenic condition, and the clinical phenotype reflects both genetic and environmental effects, making it far more challenging to define subgroups using molecular testing.128 One strategy for precision medicine in T2DM is to divide patients into subgroups based on treatment response and then examine the biological underpinnings of each subgroup utilizing next-generation sequencing platforms and gene arrays.129 Big data, or the growing availability of genetic and electronic health data from large populations, is a significant tool for delivering precision treatment for T2DM.130,131
Conclusion
The increasing incidence of diabetes is causing rising health-care costs, morbidity, mortality, and diabetes-related comorbidities. Numerous genomic technologies have led to the identification of several genetic loci associated with T2DM. However, the complete landscape of T2DM-susceptibility gene variants remains inadequate, calling for more genetic studies on various ethnicities. Moreover, it is also imperative to replicate studies on the identified gene variants through advanced sequencing technologies on different populations and subethnic groups to establish more compelling data for clinical translation. Although genomic interventions in monogenic diabetes are translated into clinical practice, they are still evolving in complex polygenic diseases like T2DM. Paradigm shifts in the future of diabetes management are crucial in tackling the diabetes epidemic. With diverse phenotypic and genotypic features in T2DM populations, the “one size fits all” approach is inept. Comprehensive phenotyping and genotyping of diabetic individuals at the prediabetic stage helps in precision diagnostics, prevention, prognostics, and therapy. Health-care professionals can use electronic medical records consisting of individuals complete omics data, including genomics, proteomics, metabolomics, and transcriptomics. Then, decisions on therapeutic optimization can be made using potential actionable findings generated in T2DM individuals. Given the remarkable advancements made over the recent decades, it is reasonable to forecast the acceptance of “personalized diabetology” in T2DM in coming years. Recent breakthroughs in genetic techniques, the application of candidate-gene studies, large-scale genotyping investigations, and GWASs have begun to produce suggestive results that may lead to changes in clinical practice. Pharmacogenetic research has already begun to deliver on the promise of personalized diabetes treatment for some monogenic forms. The recently introduced “miRNA pharmacogenomics,” which examines polymorphisms in the miRNA regulatory pathway and their relationship to drug response, would also be valuable for personalized medicine.
Author Contributions
All authors made a significant contribution to the work reported, such as conception and design, acquisition of data, analysis and interpretation of data, and drafting the article or revising it critically for important intellectual content, have agreed to submit to the current journal, gave final approval to the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ali O. Genetics of type 2 diabetes. World J Diabetes. 2013;4(4):114–123. doi:10.4239/wjd.v4.i4.114
2. Gujral UP, Pradeepa R, Weber MB, Narayan KMV, Mohan V. Type 2 diabetes in South Asians: similarities and differences with white Caucasian and other populations. Ann NY Acad Sc. 2013;1281(1):51–63. doi:10.1111/j.1749-6632.2012.06838.x
3. International Diabetes Federation. IDF Diabetes Atlas, 9th edn. Brussels, Belgium; 2019. Available from: https://www.diabetesatlas.org.
4. Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(3):1047–1053. doi:10.2337/diacare.27.5.1047
5. Joshi SR, Das AK, Vijay VJ, Mohan V. Challenges in diabetes care in India: sheer numbers, lack of awareness and inadequate control. J Assoc Physicians India. 2008;56:443–450.
6. Marín-Peñalver JJ, Martín-Timón I, Sevillano-Collantes C, Javier Del Cañizo-Gómez F. Update on the treatment of type 2 diabetes mellitus. World J Diabetes. 2016;7(17):354–395. doi:10.4239/wjd.v7.i17.354
7. Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi:10.1038/ng.120
8. Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–989. doi:10.1038/ng.921
9. Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi:10.1038/ng.609
10. Radha V, Vimaleswaran KS, Ayyappa KA, Mohan V. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int J Obes. 2007;31(6):913–918. doi:10.1038/sj.ijo.0803547
11. Vimaleswaran KS, Radha V, Ramya K, et al. A novel association of a polymorphism in the first intron of adiponectin gene with type 2 diabetes, obesity and hypoadiponectinemia in Asian Indians. Hum Genet. 2008;123(6):599–605. doi:10.1007/s00439-008-0506-8
12. Been LF, Hatfield JL, Shankar A, et al. A low frequency variant within the GWAS locus of MTNR1B affects fasting glucose concentrations: genetic risk is modulated by obesity. Nutr Metab Cardiovasc Dis. 2012;22(11):944–951. doi:10.1016/j.numecd.2011.01.006
13. Tabassum R, Mahajan A, Chauhan G, et al. Evaluation of DOK5 as a susceptibility gene for type 2 diabetes and obesity in North Indian population. BMC Med Genet. 2010;11:35. doi:10.1186/1471-2350-11-35
14. Mahajan A, Tabassum R, Srechavali E, et al. Obesity-dependent association of TNF-LTA locus with type 2 diabetes in North Indians. J Mol Med. 2010;88(5):515–522. doi:10.1007/s00109-010-0594-5
15. Ramya K, Radha V, Ghosh S, Majumder PP, Mohan V. Genetic variations in the FTO gene are associated with type 2 diabetes and obesity in South Indians (CURES-79). Diabetes Technol Ther. 2011;13(1):33–42. doi:10.1089/dia.2010.0071
16. Chauhan G, Kaur I, Tabassum R, et al. Common variants of homocysteine metabolism pathway genes and risk of type 2 diabetes and related traits in Indians. Exp Diabetes Res. 2012;2012:960318. doi:10.1155/2012/960318
17. Prakash Dwivedi O, Tabassum R, Chauhan G, et al. Strong influence of variants near MC4R on adiposity in children and adults: a cross-sectional study in Indian population. J Hum Genet. 2013;58(1):27–32. doi:10.1038/jhg.2012.129
18. Yajnik CS, Janipalli CS, Bhaskar S, et al. FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians. Diabetologia. 2009;52(2):247–252. doi:10.1007/s00125-008-1186-6
19. Cassell PG, Neverova M, Janmohamed S, et al. An uncoupling protein 2 gene variant is associated with a raised body mass index but not Type II diabetes. Diabetologia. 1999;42(6):688–692. doi:10.1007/s001250051216
20. Taylor AE, Sandeep MN, Janipalli CS, et al. Associations of FTO and MC4R variants with obesity traits in Indians and the role of rural/urban environment as a possible effect modifier. J Obes. 2011;2011:307542. doi:10.1155/2011/307542
21. Gupta V, Vinay DG, Sovio U, et al. Association study of 25 type 2 diabetes related loci with measures of obesity in Indian Sib Pairs. PLoS One. 2013;8(1):e53944. doi:10.1371/journal.pone.0053944
22. Rees SD, Hydrie MZI, O’Hare JP, et al. Effects of 16 genetic variants on fasting glucose and type 2 diabetes in South Asians: ADCY5 and GLIS3 variants may predispose to type 2 diabetes. PLoS One. 2011;6(9):e24710. doi:10.1371/journal.pone.0024710
23. Chandak GR, Janipalli CS, Bhaskar S, et al. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50(1):63–67. doi:10.1007/s00125-006-0502-2
24. Ali S, Chopra R, Manvati S, et al. Replication of type 2 diabetes candidate genes variations in three geographically unrelated Indian population groups. PLoS One. 2013;8(3):e58881. doi:10.1371/journal.pone.0058881
25. Salman M, Dasgupta S, Cholendra A, et al. MTNR1B gene polymorphisms and susceptibility to Type 2 Diabetes: a pilot study in South Indians. Gene. 2015;566(2):189–193. doi:10.1016/j.gene.2015.04.064
26. Bhatti GK, Kaur S, Vijayvergiya R, et al. ENPP1 K121Q functional variant enhances susceptibility to insulin resistance and dyslipidemia with metabolic syndrome in Asian Indians. Int J Diabetes Metab. 2018;24:8–15. doi:10.1159/000492478
27. Chavali S, Mahajan A, Tabassum R, et al. Association of variants in genes involved in pancreatic Β-cell development and function with type 2 diabetes in North Indians. J Hum Genet. 2011;56(10):695–700. doi:10.1038/jhg.2011.83
28. Chauhan G, Spurgeon CJ, Tabassum R, et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes. 2010;59(8):2068–2074. doi:10.2337/db09-1386
29. Srivastava N, Prakash J, Lakhan R, Agarwal CG, Pant DC, Mittal B. A common polymorphism in the promoter of UCP2 is associated with obesity and hyperinsulenemia in northern Indians. Mol Cell Biochem. 2010;337(1–2):293–298. doi:10.1007/s11010-009-0311-2
30. Gupta A, Gupta V, Singh AK, et al. Interleukin-6 G-174C gene polymorphism and serum resistin levels in North Indian women: potential risk of metabolic syndrome. Hum Exp Toxicol. 2011;30(10):1445–1453. doi:10.1177/0960327110393763
31. Vimaleswaran KS, Radha V, Anjana M, et al. Effect of polymorphisms in the PPARGC1A gene on body fat in Asian Indians. Int J Obes. 2006;30(6):884–891. doi:10.1038/sj.ijo.0803228
32. Gomathi P, Samarth AP, Raj NBAJ, et al. The −866G/A polymorphism in the promoter of the UCP2 gene is associated with risk for type 2 diabetes and with decreased insulin levels. Gene. 2019;701:125–130. doi:10.1016/j.gene.2019.03.041
33. Stančáková A, Laakso M. Genetics of type 2 diabetes. Endocr Dev. 2016;31:203–220. doi:10.1159/000439418
34. Prasad RB, Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes. 2015;6(1):87–123. doi:10.3390/genes6010087
35. Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20(3):284–287. doi:10.1038/3099
36. Schwanstecher C, Schwanstecher M. Nucleotide sensitivity of pancreatic ATP-sensitive potassium channels and type 2 diabetes. Diabetes. 2002;51(Suppl 3):S358–S362. doi:10.2337/diabetes.51.2007.s358
37. Hani EH, Boutin P, Durand E, et al. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia. 1998;41:1511–1515. doi:10.1007/s001250051098
38. Gloyn AL, Pearson ER, Antcliff JF, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. doi:10.1056/nejmoa032922
39. Almind K, Bj o Rbaek C, Vestergaard H, Hansen T, Echwald S, Pedersen O. Aminoacid polymorphisms of insulin receptor substrate-1 in non-insulin-dependent diabetes mellitus. Lancet. 1993;342(8875):828–832. doi:10.1016/0140-6736(93)92694-O
40. Le Fur S, Le Stunff C, Bougnères P. Increased insulin resistance in obese children who have both 972 IRS-1 and 1057 IRS-2 polymorphisms. Diabetes. 2002;51(Suppl 3):S304–S307. doi:10.2337/diabetes.51.2007.s304
41. Cheng S, Wu Y, Wu W, Zhang D. Association of rs734312 and rs10010131 polymorphisms in WFS1 gene with type 2 diabetes mellitus: a meta-analysis. Endocr J. 2013;60(4):441–447. doi:10.1507/endocrj.ej12-0325
42. Giuffrida FMA, Furuzawa GK, Kasamatsu TS, Oliveira MM, Reis AF, Dib SA. HNF1A gene polymorphisms and cardiovascular risk factors in individuals with late-onset autosomal dominant diabetes: a cross-sectional study. Cardiovasc Diabetol. 2009;8:28. doi:10.1186/1475-2840-8-28
43. Abate N, Chandalia M, Satija P, et al. ENPP1/PC-1 K121Q polymorphism and genetic susceptibility to type 2 diabetes. Diabetes. 2005;54(4):1207–1213. doi:10.2337/diabetes.54.4.1207
44. Moore AF, Jablonski KA, Mason CC, et al. The association of ENPP1 K121Q with diabetes incidence is abolished by lifestyle modification in the diabetes prevention program. J Clin Endocrinol Metab. 2009;94(2):449–455. doi:10.1210/jc.2008-1583
45. Li YY. ENPP1 K121Q polymorphism and type 2 diabetes mellitus in the Chinese population: a meta-analysis including 11 855 subjects. Metabolism. 2012;61(5):625–633. doi:10.1016/j.metabol.2011.10.002
46. Bhatti JS, Bhatti GK, Mastana SS, Ralhan S, Joshi A, Tewari R. ENPP1/PC-1 K121Q polymorphism and genetic susceptibility to type 2 diabetes in North Indians. Mol Cell Biochem. 2010;345(1–2):249–257. doi:10.1007/s11010-010-0579-2
47. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi:10.1038/nature03001
48. International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi:10.1038/nature02168
49. Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi:10.1038/nature05616
50. Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi:10.1038/ng.2982
51. Smith RJ, Nathan DM, Arslanian SA, Groop L, Rizza RA, Rotter JI. Individualizing therapies in type 2 diabetes mellitus based on patient characteristics: what we know and what we need to know. J Clin Endocrinol Metab. 2010;95(4):1566–1574. doi:10.1210/jc.2009-1966
52. Gillies CL, Abrams KR, Lambert PC, et al. Pharmacological and lifestyle interventions to prevent or delay type 2 diabetes in people with impaired glucose tolerance: systematic review and meta-analysis. Br Med J. 2007;334(7588):299–302. doi:10.1136/bmj.39063.689375.55
53. Ahlqvist E, Storm P, Käräjämäki A, et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6(5):361–369. doi:10.1016/S2213-8587(18)30051-2
54. Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. A primer on deep learning in genomics. Nat Genet. 2019;51(1):12–18. doi:10.1038/s41588-018-0295-5
55. Ge S, Wang Y, Song M, et al. Type 2 diabetes mellitus: integrative analysis of multiomics data for biomarker discovery. OMICS. 2018;22(7):514–523. doi:10.1089/omi.2018.0053
56. Van Der Heijden AA, Rauh SP, Dekker JM, et al. The Hoorn Diabetes Care System (DCS) cohort. A prospective cohort of persons with type 2 diabetes treated in primary care in the Netherlands. BMJ Open. 2017;7(5):e015599. doi:10.1136/bmjopen-2016-015599
57. Andrew H, Neil W, Demicco DA, et al. Analysis of efficacy and safety in patients aged 65–75 years at randomization collaborative atorvastatin diabetes study (CARDS). Diabetes Care. 2006;29(11):2378–2384. doi:10.2337/dc06-0872
58. Ikram MA, Brusselle GGO, Murad SD, et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur J Epidemiol. 2017;32(9):807–850. doi:10.1007/s10654-017-0321-4
59. Van Leeuwen N, Nijpels G, Becker ML, et al. A gene variant near ATM is significantly associated with metformin treatment response In type 2 diabetes: a replication and meta-analysis of five cohorts. Diabetologia. 2012;55(7):1971–1977. doi:10.1007/s00125-012-2537-x
60. Shokri F, Ghaedi H, Ghafouri Fard S, et al. Impact of ATM and SLC22A1 polymorphisms on therapeutic response to metformin in Iranian diabetic patients. Int J Mol Cell Med. 2016;5(1):1–7.
61. Out M, Becker ML, Van Schaik RH, Lehert P, Stehouwer CD, Kooy A. A gene variant near ATM affects the response to metformin and metformin plasma levels: a post hoc analysis of an RCT. Pharmacogenomics. 2018;19(8):715–726. doi:10.2217/pgs-2018-0010
62. Vilvanathan S, Gurusamy U, Mukta V, Das AK, Chandrasekaran A. Allele and genotype frequency of a genetic variant in ataxia telangiectasia mutated gene affecting glycemic response to metformin in South Indian population. Indian J Endocrinol Metab. 2014;18(6):850–854. doi:10.4103/2230-8210.119944
63. Umamaheswaran G, Praveen RG, Damodaran SE, Das AK, Adithan C. Influence of SLC22A1 rs622342 genetic polymorphism on metformin response in South Indian type 2 diabetes mellitus patients. Clin Exp Med. 2015;15(4):511–517. doi:10.1007/s10238-014-0322-5
64. Dujic T, Zhou K, Donnelly LA, Tavendale R, Palmer CNA, Pearson ER. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: a GoDARTS study. Diabetes. 2015;64(5):1786–1793. doi:10.2337/db14-1388
65. Sur D. A tale of genetic variation in the human slc22a1 gene encoding oct1 among type 2 diabetes mellitus population groups of West Bengal, India. IMPACT. 2014;2:97–106. doi:10.4172/1747-0862.C1.010
66. Mato EPM, Guewo-Fokeng M, Faadiel Essop M, Owira PMO. Genetic polymorphisms of organic cation transporters 1 (OCT1) and responses to metformin therapy in individuals with type 2 diabetes mellitus: a systematic review. Medicine. 2018;97(27):e11349. doi:10.1097/MD.0000000000011349
67. Reséndiz-Abarca CA, Flores-Alfaro E, Suárez-Sánchez F, et al. Altered glycemic control associated with polymorphisms in the SLC22A1 (OCT1) gene in a Mexican population with type 2 diabetes mellitus treated with metformin: a cohort study. J Clin Pharmacol. 2019;59(10):1384–1390. doi:10.1002/jcph.1425
68. Todd JN, Florez JC. An update on the pharmacogenomics of metformin: progress, problems and potential. Pharmacogenomics. 2014;15(4):529–539. doi:10.2217/pgs.14.21
69. Becker ML, Visser LE, Van Schaik RHN, Hofman A, Uitterlinden AG, Stricker BHC. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58(3):745–749. doi:10.2337/db08-1028
70. Tzvetkov MV, Vormfelde SV, Balen D, et al. The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin Pharmacol Ther. 2009;86(3):299–306. doi:10.1038/clpt.2009.92
71. Al-Eitan LN, Almomani BA, Nassar AM, Elsaqa BZ, Saadeh NA. Metformin pharmacogenetics: effects of SLC22A1, SLC22A2, and SLC22A3 polymorphisms on glycemic control and HBA1c levels. J Pers Med. 2019;9(1):17. doi:10.3390/jpm9010017
72. Ghaffari-Cherati M, Mahrooz A, Hashemi-Soteh MB, Raheleh Hosseyni-Talei S, Alizadeh A, Nakhaei SM. Allele frequency and genotype distribution of a common variant in the 3´-untranslated region of the SLC22A3 gene in patients with type 2 diabetes: association with response to metformin. J Res Med Sci. 2016;21:92. doi:10.4103/1735-1995.192508
73. Rotroff DM, Yee SW, Zhou K, et al. Genetic variants in CPA6 and PRPF31 are associated with variation in response to metformin in individuals with type 2 diabetes. Diabetes. 2018;67(7):1428–1440. doi:10.2337/db17-1164
74. Dujic T, Bego T, Malenica M, et al. Effects of TCF7L2 rs7903146 variant on metformin response in patients with type 2 diabetes. Bosn J Basic Med Sci. 2019;19(4):368–374. doi:10.17305/bjbms.2019.4181
75. Dawed AY, Zhou K, van Leeuwen N, et al. Variation in the plasma membrane monoamine transporter (PMAT) (encoded by SLC29A4) and organic cation transporter 1 (OCT1) (encoded by SLC22A1) and gastrointestinal intolerance to metformin in type 2 diabetes: an IMI DIRECT study. Diabetes Care. 2019;42(6):1027–1033. doi:10.2337/dc18-2182
76. Becker ML, Visser LE, Trienekens PH, Hofman A, Van Schaik RHN, Stricker BHC. Cytochrome P450 2C9 *2 and *3 polymorphisms and the dose and effect of sulfonylurea in type II diabetes mellitus. Clin Pharmacol Ther. 2008;83(2):288–292. doi:10.1038/sj.clpt.6100273
77. Weisnagel SJ, Rankinen T, Nadeau A, et al. Decreased fasting and oral glucose stimulated C-peptide in nondiabetic subjects with sequence variants in the sulfonylurea receptor 1 gene. Diabetes. 2001;50(3):697–702. doi:10.2337/diabetes.50.3.697
78. Venkatesan R, Bodhini D, Narayani N, Mohan V. Association study of the ABCC8 gene variants with type 2 diabetes in south Indians. Indian J Hum Genet. 2014;20(1):37–42. doi:10.4103/0971-6866.132752
79. Javorsky M, Klimcakova L, Schroner Z, et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med. 2012;23(3):245–249. doi:10.1016/j.ejim.2011.10.018
80. Loganadan NK, Huri HZ, Vethakkan SR, Hussein Z. Genetic markers predicting sulphonylurea treatment outcomes in type 2 diabetes patients: current evidence and challenges for clinical implementation. Pharmacogenomics J. 2016;16(3):209–219. doi:10.1038/tpj.2015.95
81. Dhawan D, Padh H. Genetic variations in TCF7L2 influence therapeutic response to sulfonylureas in Indian diabetics. Diabetes Res Clin Pract. 2016;121:35–40. doi:10.1016/j.diabres.2016.08.018
82. Schroner Z, Javorsky M, Tkacova R, et al. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes Metab. 2011;13(11):89–91. doi:10.1111/j.1463-1326.2010.01324.x
83. Holstein A, Hahn M, Körner A, Stumvoll M, Kovacs P. TCF7L2 and therapeutic response to sulfonylureas in patients with type 2 diabetes. BMC Med Genet. 2011;12:30. doi:10.1186/1471-2350-12-30
84. Chen ZR, He FZ, Liu MZ, et al. MIR4532 gene variant rs60432575 influences the expression of KCNJ11 and the sulfonylureas-stimulated insulin secretion. Endocrine. 2019;63(3):489–496. doi:10.1007/s12020-018-1754-6
85. Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27(4):740–756. doi:10.1016/j.cmet.2018.03.001
86. Buse JB, Wexler DJ, Tsapas A, et al. 2019 update to: management of hyperglycemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care. 2020;43(2):487–493. doi:10.2337/dci19-0066
87. American Diabetes Association. 9. Pharmacologic approaches to glycemic treatment: standards of medical care in diabetes-2020. Diabetes Care. 2020;43(Suppl1):S98–S110. doi:10.2337/dc20-S009
88. Nauck M. Incretin therapies: highlighting common features and differences in the modes of action of glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors. Diabetes Obes Metab. 2016;18(3):203–216. doi:10.1111/dom.12591
89. Urgeová A, Javorský M, Klimčáková L, et al. Genetic variants associated with glycemic response to treatment with dipeptidylpeptidase 4 inhibitors. Pharmacogenomics. 2020;21(5):317–323. doi:10.2217/pgs-2019-0147
90. Böhm A, Wagner R, Machicao F, et al. DPP4 gene variation affects GLP-1 secretion, insulin secretion, and glucose tolerance in humans with high body adiposity. PLoS One. 2017;12(7):e0181880. doi:10.1371/journal.pone.0181880
91. Jamaluddin JL, Huri HZ, Vethakkan SR. Clinical and genetic predictors of dipeptidyl peptidase-4 inhibitor treatment response in Type 2 diabetes mellitus. Pharmacogenomics. 2016;17(8):867–881. doi:10.2217/pgs-2016-0010
92. Gotthardová I, Javorský M, Klimčáková L, et al. KCNQ1 gene polymorphism is associated with glycaemic response to treatment with DPP-4 inhibitors. Diabetes Res Clin Pract. 2017;130:142–147. doi:10.1016/j.diabres.2017.05.018
93. Osada UN, Sunagawa H, Terauchi Y, Ueda S. A common susceptibility gene for type 2 diabetes is associated with drug response to a DPP-4 inhibitor: pharmacogenomic cohort in Okinawa Japan. PLoS One. 2016;11(5):e0154821. doi:10.1371/journal.pone.0154821
94. Liao WL, Lee WJ, Chen CC, et al. Pharmacogenetics of dipeptidyl peptidase 4 inhibitors in a Taiwanese population with type 2 diabetes. Oncotarget. 2017;8(11):18050–18058. doi:10.18632/oncotarget.14951
95. Zimdahl H, Haupt A, Brendel M, et al. Influence of common polymorphisms in the SLC5A2 gene on metabolic traits in subjects at increased risk of diabetes and on response to empagliflozin treatment in patients with diabetes. Pharmacogenet Genomics. 2017;27(4):135–142. doi:10.1097/FPC.0000000000000268
96. Enigk U, Breitfeld J, Schleinitz D, et al. Role of genetic variation in the human sodium-glucose cotransporter 2 gene (SGLT2) in glucose homeostasis. Pharmacogenomics. 2011;12(8):1119–1126. doi:10.2217/pgs.11.69
97. Francke S, Mamidi RNVS, Solanki B, et al. In vitro metabolism of canagliflozin in human liver, kidney, intestine microsomes, and recombinant uridine diphosphate glucuronosyltransferases (UGT) and the effect of genetic variability of UGT enzymes on the pharmacokinetics of canagliflozin in humans. J Clin Pharmacol. 2015;55(9):1061–1072. doi:10.1002/jcph.506
98. Chiasson JL, Gomis R, Hanefeld M, Josse RG, Karasik A, Laakso M. The STOP-NIDDM trial: an international study on the efficacy of an α- glucosidase inhibitor to prevent type 2 diabetes in a population with impaired glucose tolerance: rationale, design, and preliminary screening data. Diabetes Care. 1998;21(10):1720–1725. doi:10.2337/diacare.21.10.1720
99. Andrulionytè L, Zacharova J, Chiasson JL, Laakso M. Common polymorphisms of the PPAR-γ2 (Pro12Ala) and PGC-1α (Gly482Ser) genes are associated with the conversion from impaired glucose tolerance to type 2 diabetes in the STOP-NIDDM trial. Diabetologia. 2004;47(12):2176–2184. doi:10.1007/s00125-004-1577-2
100. Hart LM, Fritsche A, Nijpels G, et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes. 2013;62(9):3275–3281. doi:10.2337/db13-0227
101. Kan H, Hyogo H, Ochi H, et al. Influence of the rs738409 polymorphism in patatin-like phospholipase 3 on the treatment efficacy of non-alcoholic fatty liver disease with type 2 diabetes mellitus. Hepatol Res. 2016;46(3):E146–E153. doi:10.1111/hepr.12552
102. Chedid V, Vijayvargiya P, Carlson P, et al. Allelic variant in the glucagon-like peptide 1 receptor gene associated with greater effect of liraglutide and exenatide on gastric emptying: a pilot pharmacogenetics study. Neurogastroenterol Motil. 2018;30(7):e13313. doi:10.1111/nmo.13313
103. Lin CH, Lee YS, Huang YY, Hsieh SH, Chen ZS, Tsai CN. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res. 2015;2015:176949. doi:10.1155/2015/176949
104. Singh S, Usman K, Banerjee M. Pharmacogenetic studies update in type 2 diabetes mellitus. World J Diabetes. 2016;7(15):302–315. doi:10.4239/wjd.v7.i15.302
105. Niemi M, Leathart JB, Neuvonen M, Backman JT, Daly AK, Neuvonen PJ. Polymorphism in CYP2C8 is associated with reduced plasma concentrations of repaglinide. Clin Pharmacol Ther. 2003;74(4):380–387. doi:10.1016/S0009-9236(03)00228-5
106. Sheng FF, Dai XP, Qu J, et al. NAMPT −3186C/T polymorphism affects repaglinide response in Chinese patients with Type 2 diabetes mellitus. Clin Exp Pharmacol Physiol. 2011;38(8):550–554. doi:10.1111/j.1440-1681.2011.05548.x
107. Dai XP, Huang Q, Yin JY, et al. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese Type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2012;39(5):462–468. doi:10.1111/j.1440-1681.2012.05701.x
108. Wang T, Wang Y, Lv DM, et al. Effects of NOS1AP rs12742393 polymorphism on repaglinide response in Chinese patients with type 2 diabetes mellitus. Pharmacotherapy. 2014;34(2):131–139. doi:10.1002/phar.1379
109. Zhou X, Chen C, Yin D, et al. A variation in the ABCC8 gene is associated with type 2 diabetes mellitus and repaglinide efficacy in Chinese type 2 diabetes mellitus patients. Intern Med. 2019;58(16):2341–2347. doi:10.2169/internalmedicine.2133-18
110. Quinn CE, Hamilton PK, Lockhart CJ, McVeigh GE. Thiazolidinediones: effects on insulin resistance and the cardiovascular system. Br J Pharmacol. 2008;153(4):636–645. doi:10.1038/sj.bjp.0707452
111. Chang TJ, Liu PH, Liang YC, et al. Genetic predisposition and nongenetic risk factors of thiazolidinedione-related edema in patients with type 2 diabetes. Pharmacogenet Genomics. 2011;21(12):829–836. doi:10.1097/FPC.0b013e32834bfff1
112. Zhang KH, Huang Q, Dai XP, et al. Effects of the peroxisome proliferator activated receptor-γ coactivator-1α (PGC-1α) Thr394Thr and Gly482Ser polymorphisms on rosiglitazone response in Chinese patients with type 2 diabetes mellitus. J Clin Pharmacol. 2010;50(9):1022–1030. doi:10.1177/0091270009355159
113. Eun SK, So YP, Hyeong JK, et al. Effects of Pro12Ala polymorphism of peroxisome proliferator-activated receptor γ2 gene on rosiglitazone response in type 2 diabetes. Clin Pharmacol Ther. 2005;78(2):202–208. doi:10.1016/j.clpt.2005.04.013
114. Hsieh MC, Der LK, Tien KJ, et al. Common polymorphisms of the peroxisome proliferator-activated receptor-γ (Pro12Ala) and peroxisome proliferator-activated receptor-γ coactivator-1 (Gly482Ser) and the response to pioglitazone in Chinese patients with type 2 diabetes mellitus. Metabolism. 2010;59(8):1139–1144. doi:10.1016/j.metabol.2009.10.030
115. Chen M, Hu C, Zhang R, et al. Association of PAX4 genetic variants with oral antidiabetic drugs efficacy in Chinese type 2 diabetes patients. Pharmacogenomics J. 2014;14(5):488–492. doi:10.1038/tpj.2014.18
116. Dawed AY, Donnelly L, Tavendale R, et al. CYP2C8 and SLCO1B1 variants and therapeutic response to thiazolidinediones in patients with Type 2 diabetes. Diabetes Care. 2016;39(11):1902–1908. doi:10.2337/dc15-2464
117. Hruska MW, Amico JA, Langaee TY, Ferrell RE, Fitzgerald SM, Frye RF. The effect of trimethoprim on CYP2C8 mediated rosiglitazone metabolism in human liver microsomes and healthy subjects. Br J Clin Pharmacol. 2005;59(1):70–79. doi:10.1111/j.1365-2125.2005.02263.x
118. Chung WK, Erion K, Florez JC, et al. Precision medicine in diabetes: a consensus report from the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia. 2020;63(9):1671–1693. doi:10.1007/s00125-020-05181-w
119. Della Corte CM, Ciaramella V, Di Mauro C, et al. Metformin increases antitumor activity of MEK inhibitors through GLI1 downregulation in LKB1 positive human NSCLC cancer cells. Oncotarget. 2016;7(4):4265–4278. doi:10.18632/oncotarget.6559
120. Morgillo F, Fasano M, Della Corte CM, et al. Results of the safety run-in part of the METAL (METformin in Advanced Lung cancer) study: a multicentre, open-label Phase I-II study of metformin with erlotinib in second-line therapy of patients with stage IV non-small-cell lung cancer. ESMO Open. 2017;2(2):e000132. doi:10.1136/esmoopen-2016-000132
121. Sasso FC, Pafundi PC, Simeon V, et al. Efficacy and durability of multifactorial intervention on mortality and MACEs: a randomized clinical trial in type-2 diabetic kidney disease. Cardiovasc Diabetol. 2021;20(1):145. doi:10.1186/s12933-021-01343-1
122. Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–677. doi:10.1007/s00439-017-1779-6
123. Landrum MJ, Chitipiralla S, Brown GR, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–844. doi:10.1093/nar/gkz972
124. Patel DK, Strong J. The pleiotropic effects of sodium-glucose cotransporter-2 inhibitors: beyond the glycemic benefit. Diabetes Ther. 2019;10(5):1771–1792. doi:10.1007/s13300-019-00686-z
125. Shao SC, Chang KC, Lin SJ, et al. Favorable pleiotropic effects of sodium glucose cotransporter 2 inhibitors: head-to-head comparisons with dipeptidyl peptidase-4 inhibitors in type 2 diabetes patients. Cardiovasc Diabetol. 2020;19(1):17. doi:10.1186/s12933-020-0990-2
126. Tang L, Chang SJ, Chen CJ, Liu JT. Non-invasive blood glucose monitoring technology: a review. Sensors. 2020;20(23):1–32. doi:10.3390/s20236925
127. Sirdah MM, Reading NS. Genetic predisposition in type 2 diabetes: a promising approach toward a personalized management of diabetes. Clin Genet. 2020;98(6):525–547. doi:10.1111/cge.13772
128. Florez JC. Pharmacogenetics in type 2 diabetes: precision medicine or discovery tool? Diabetologia. 2017;60(5):800–807. doi:10.1007/s00125-017-4227-1
129. Huang C, Florez JC. Pharmacogenetics in type 2 diabetes: potential implications for clinical practice. Genome. 2011;3(76). doi:10.1186/gm292
130. Klonoff DC. Personalized medicine for diabetes. J Diabetes Sci Technol. 2008;2(3):335–341. doi:10.1177/193229680800200301
131. Maruthur NM, Gribble MO, Bennett WL, et al. The pharmacogenetics of type 2 diabetes: a systematic review. Diabetes Care. 2014;37(3):876–886. doi:10.2337/dc13-1276
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.