Back to Journals » International Journal of Nanomedicine » Volume 17
Peripheral Serum Exosomes Isolated from Patients with Acute Myocardial Infarction Promote Endothelial Cell Angiogenesis via the miR-126-3p/TSC1/mTORC1/HIF-1α Pathway
Authors Duan S , Wang C, Xu X, Zhang X, Su G, Li Y , Fu S, Sun P, Tian J
Received 12 September 2021
Accepted for publication 7 March 2022
Published 2 April 2022 Volume 2022:17 Pages 1577—1592
DOI https://doi.org/10.2147/IJN.S338937
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Shasha Duan,1– 3,* Chao Wang,1,2,* Xiangli Xu,4 Xiaoshan Zhang,3 Gaofeng Su,1,2 You Li,1,2 Shuai Fu,1,2 Ping Sun,1,2 Jiawei Tian1
1Department of Ultrasound, the Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang Province, People’s Republic of China; 2The Key Laboratory of Myocardial Ischemia, Harbin Medical University, Ministry of Education, Harbin, Heilongjiang Province, People’s Republic of China; 3Department of Ultrasound, The Affiliated Hospital of Inner Mongolia Medical University, Hohhot, People’s Republic of China; 4Department of Ultrasound, the Second Hospital of Harbin city, Harbin, Heilongjiang Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiawei Tian; Ping Sun, Department of Ultrasound, the Second Affiliated Hospital of Harbin Medical University, No. 246 Xuefu Road, Nangang District, Harbin, Heilongjiang Province, 150086, People’s Republic of China, Tel +86 451-86605811, Fax +86 451-86605745, Email [email protected]; [email protected]
Purpose: Angiogenesis is required for improving myocardial function and is a key factor in long-term prognosis after an acute myocardial infarction (AMI). Although exosomes are known to play a crucial role in angiogenesis, the role of peripheral exosomes in angiogenic signal transduction in patients with AMI remains unclear. Here, we explored the effect of exosomes extracted from the peripheral serum of AMI patients on angiogenesis and elucidated the downstream pathways.
Patients and Methods: Serum exosomes were obtained from patients with AMI (AMI-Exo) and healthy individuals (Con-Exo). The exosomes were cocultured with human umbilical vein endothelial cells (HUVECs) in vitro, with aortic rings ex vivo, and were used to treat mouse hind-limb ischemia and mouse AMI model in vivo.
Results: AMI-Exo raised HUVEC proliferation, tube formation, and migration, and enhanced microvessel sprouting from aortic rings compared to Con-Exo, both in vitro and ex vivo. Quantitative reverse transcription-polymerase chain reaction revealed that the abundance of miR-126-3p, a crucial regulator of angiogenesis, was increased in AMI-Exo. The inhibition of miR-126-3p decreased the benefits of AMI-Exo treatment, and miR-126-3p upregulation enhanced the benefits of Con-Exo treatment in HUVECs, aortic rings, the mouse hind-limb ischemia model, and the mouse AMI model. Knockdown and overexpression analyses revealed that miR-126-3p regulated angiogenesis in HUVECs by directly targeting tuberous sclerosis complex 1 (TSC1). Moreover, we found that miR-126-3p could inhibit TSC1 expression, which further activated mTORC1 signaling and increased HIF-1α and VEGFA expression, ultimately promoting angiogenesis.
Conclusion: Collectively, our results provide a novel understanding of the function of exosomes in angiogenesis post AMI. We demonstrated that exosomes from the peripheral serum of AMI patients promote angiogenesis via the miR-126-3p/TSC1/mTORC1/HIF-1α signaling pathway.
Keywords: acute myocardial infarction, angiogenesis, exosomes, miR-126-3p, tuberous sclerosis complex 1
Introduction
Cardiovascular diseases (CVDs) pose immense health and economic burdens worldwide, and account for 44% of all disease-related deaths in China.1 Acute myocardial infarction (AMI) is the leading cause of death from CVDs. Although thrombolytic anticoagulant therapy and percutaneous coronary intervention can largely restore occluded coronary perfusion after AMI, they cannot repair the damage caused to microvascular architecture in ischemic myocardium by coronary hypoxic ischemia. Angiogenesis is a complex process of vascular endothelial cell proliferation, migration, and branching that forms new microvessels from existing vessels in response to various factors. Since angiogenesis plays an important role in improving myocardial function after AMI,2 it is necessary to explore the underlying mechanisms and identify effective new therapeutic targets.
Exosomes are a type of extracellular microvesicle, with diameters ranging from 30 nm to 150 nm. They contain all kinds of functional biomolecules (DNA, mRNAs, lipids, proteins, and microRNAs [miRNAs]) that can be transferred to nearby or distant cells via the circulatory system.3 Previous studies have shown that plasma exosomes derived from adult rats and humans can protect the myocardium from ischemia-reperfusion injury by delivering endogenous protective signals to the myocardium.4 In addition, it has been reported that cardiomyocytes from mice with ischemic injury may release cell-dependent exosomes into the circulatory system.5 During transport, exosomes can protect bioactive molecules from degradation.6 For instance, Cao et al7 reported that during AMI convalescence, circulating exosomes can be transported between cells through dynamin, and transport miR-193a-5p to protect endothelial cells experiencing oxidative stress. However, it remains unclear whether exosomes derived from the circulatory serum during early phases of AMI can play regulatory roles in endothelial cell angiogenesis.
In this study, we researched the function of circulating exosomes in angiogenesis; the exosomes were isolated from AMI patients within 12 h of clinical symptom onset (AMI-Exo) and healthy individuals (Con-Exo). The function of human umbilical vein endothelial cell (HUVEC), microvessel sprouting from aortic rings in ex vivo, and the hind limb ischemia as well as AMI mouse model were studied. Taken together, our findings can provide new targets for research into the mechanisms of angiogenesis during myocardial infarction.
Patients and Methods
Patients
The diagnostic criteria for AMI patients were according to the 2012 ESC/AHA/ACC guidelines.8 The study participants were patients with suspected AMI based on electrocardiographic diagonal evidence, chest pain, and diagnostic cardiac catheterization at the Second Affiliated Hospital of Harbin Medical University between May 2018 and July 2019. The exclusion criteria were: 1) diabetes, 2) infectious diseases, and 3) other contraindications including tumors and nephritic or hepatic diseases. According to their angiography results, patients with more than 70% stenosis of the coronary artery were treated as the AMI group. Healthy individuals served as the control group. All individuals signed informed consent and the experiments were approved by the Second Affiliated Hospital of Harbin Medical University Ethics Committee (Harbin City, Heilongjiang Province, China). This experiment was conducted in accordance with the Declaration of Helsinki.
Serum Exosome Preparation and Identification
Blood samples (10 mL) were collected from the peripheral veins of all research subjects. From the AMI patients, the blood samples were collected before coronary stent implantation and within 12 h of the onset of chest pain. Serum exosomes were isolated from all subjects as described previously.9 Briefly, the serum samples were centrifuged at 1000 × g for 5 min and again at 10,000 × g for 10 min. Then, a 0.1 mm pore mesh was used to filter the cells; these were then ultracentrifuged at 100,000 × g for 1h. The exosomes were washed by PBS and subjected once again to ultracentrifugation at 100,000 × g for 1 h. The derived microvesicles were resuspended in PBS for later experiments. Bicinchoninic acid (BCA) protein kit (Beyotime, Shanghai, China) was used to quantitatively evaluate the exosome concentration. The size distribution and ultrastructure of the exosomes were analyzed using nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM; H-7650; Hitachi, Japan); their Alix, CD63, and TSG101 expression were analyzed using Western blotting.
Cell Culture
HUVECs obtained from Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China, were employed. The cells were cultured using Dulbecco modified eagle medium (DMEM) (Gibco, BRL, USA), 10% newborn calf serum (Invitrogen Life Technologies, Carlsbad, California, USA), and 1% penicillin-streptomycin (Beyotime, Shanghai, China). The culture plates were placed in an environment with the following conditions: temperature: 37°C, O2: 21%, and CO2: 5%.
Experimental Animals
Wild-type male 8–9-week-old C57BL/6 mice and 6-week-old Sprague–Dawley rats were purchased from the Medical Laboratory Animal Supply Base (Heilongjiang, China). The animals were reared in accordance with the principles of animal care approved by the National Society for Medical Research and the Guideline for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources/NIH). The experiments on animals were allowed by the Research Ethics Committee of the Second Affiliated Hospital of Harbin Medical University (SYDW2020-030).
Quantitative RT-PCR Analysis
Exosomal RNA was derived from exosomes using a miRcute miRNA Isolation Kit (Tiangen Biotech, Beijing, China) and then reverse transcribed into cDNA using an miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech), based on standard protocols. Total cellular RNA was extracted by TRIzol reagent (Invitrogen) and mRNA was reverse transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Risch-Rotkreuz, Switzerland). For exosomal miRNA, the relative levels were normalized compared to those of synthetic Caenorhabditis elegans (cel)-miR-39 (5 fmol/mL; Applied Biosystems, California, USA). Meanwhile, for cellular miRNA, relative levels were normalized with U6 RNA. Cellular mRNA levels were normalized to GAPDH mRNA standards. Quantitative analysis was conducted using the 2−ΔΔCt method. The primers are shown in Table S1.
Western Blotting Analysis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate proteins; these were then transferred onto polyvinylidene difluoride membranes (Millipore, Massachusetts, USA), and incubated at 4 °C overnight together with primary antibodies against Alix (Abcam, ab186429, UK), CD63 (Abcam, ab217345), TSG101 (Abcam, ab125011), TSC1 (CST, 6935, China), HIF-1α (Abcam, ab179483), phospho-S6 S240/244 (CST, 5364), S6 (CST, 2217), VEGFA (Abcam, ab214424), and GAPDH (Bioss, Beijing, China). Secondary antibodies were purchased from (Zsbio, Beijing, China). The expression of the genes in the samples were normalized to that of GAPDH.
HUVEC Proliferation, Tube Formation, and Migration
HUVEC proliferation was evaluated using Cell Counting kit-8 (Dojindo, CK04-11, Japan), Cell-Light EdU (5-ethynyl-2′-deoxyuridine) kit (Ribobio, C10310-3, Guangzhou, China), and manual cell counting assay. Briefly, 100 µL of cell suspension including approximately 3000 HUVECs was placed in a 96-well plate and treated with 200 μg/mL AMI-Exo, Con-Exo, and PBS, respectively. After 24 h, CCK-8 reagent (10 μL) was added and the cells were cultivated for 3 h. Then, spectrophotometric microplate reader was used for measuring the optical density (OD) at 450 nm. In the EdU assay, the labelling reagent was added to a 96-well plate and the HUVECs were allowed to proliferate for 2 h. The cells were imaged using a fluorescence microscope (Leica, Germany).
HUVEC migration was analyzed using Transwell inserts (Corning, 3422, USA) and scratch wound assays. Cells were resuspended in serum-free DMEM, dispensed in the Transwell inserts (approximately 2×105 cells/well); the cells then migrated to the underside containing DMEM with 10% FBS, through the 8.0 µm pore polycarbonate membrane. The cells were then treated with PBS, Con-Exo, AMI-Exo, and cultured for 8 h at 37°C with 5% CO2. Cells within each insert were removed using cotton swabs. The migrated cells were treated with crystal violet for 10 min and quantitated in three random microscopic fields.
Scratch wound assays were carried out by resuspending HUVECs seeded in 6-well plates and starving them for 12h. A scratch wound was created using a yellow pipette tip and then, PBS, Con-Exo or AMI-Exo were added, respectively. After 24 h, the wound area was observed and analyzed by ImageJ software. The ratio of non-migrated area divided by the baseline area was used to calculate the migration effects.
In in-vitro tube formation assay, 50 μL Matrigel Matrix (BD, 356234) was added to a 96-well plate. Then, about 2×104 cells/well were seeded on the Matrigel and co-cultured with PBS, Con-Exo, or AMI-Exo for 8 h. The total tube length and number of nodes were quantitatively analyzed by ImageJ software.
Aortic Ring Assay
All procedures were performed under sterile conditions. Briefly, 1 mL Matrigel Matrix (BD, 356234) was mixed with serum-free medium 1:1 in a 2.5 mL Eppendorf (EP) tube, added to a 96-well plate (70 μL/well), and incubated at 37 °C for 30 min. Six-week old Sprague–Dawley rats were euthanized and their thoracic aorta dissected. After the removal of the surrounding tissue and fat, the aorta was cut into ~1 mm sections. One ring was gently placed in each well of a 96-well plate with 70 μL mixed Matrigel in serum-free medium 1:1, and incubated at 37 °C. Ring sprouting was visualized on day 4 using a light microscope with a phase-contrast setting. Sprout outgrowths were counted manually. The number of sprouts that emerged from the main ring as well as the individual branches arising from it as separate vessels were counted.10
Luciferase Assay
Overlap analysis revealed that TSC1 may be a potential therapeutic target. Wild-type and mutated TSC1 mRNA 3ʹ UTR fragments were synthesized and inserted into the p-MIR-REPORT plasmid (GenePharma, Shanghai, China). The 293T cells were co-transfected with 60 nM of mimic miR-126-3p or inhibitor and 10 ng of Renilla luciferase reporter. After 24 h, Renilla luciferase activity was evaluated by Dual-Luciferase Reporter Assay System (Promega, USA), based on the manufacturer’s protocol.
Statistical Analysis
Continuous variables were presented as the mean ± standard deviation or as the median and interquartile range. Student’s t-test or Mann–Whitney U-test, as appropriate. Categorical variables were expressed as numbers and percentages, and analyzed using the chi-square test. The SPSS statistical software (version 25.0; SPSS Inc., IBM Corp., Armonk, NY, USA) was employed. Two-sided P values of < 0.05 were considered as statistically significant. Histograms were drawn using GraphPad Prism 8.0 (GraphPad Software, California, USA).
Further information on the methods is described in the Supplementary Information.
Results
Serum Exosome Characterization
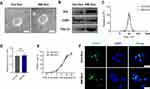
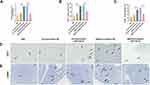
Twenty AMI patients and 10 healthy individuals were included in this study. Their clinical baseline characteristics are shown in Table 1. Serum exosomes from the healthy individuals and AMI patients were derived by ultracentrifugation, as reported previously,11 and their morphology was analyzed using TEM. The exosomes displayed a round, cup-shaped morphology (Figure 1A). Specific exosomal markers (Alix, CD63, and TSG101) were expressed in both exosome types (Figure 1B), and NanoSight measurement showed that they both displayed particle size distributions of 30–150 nm (Figure 1C). Meanwhile, there was no statistical difference in concentration between Con-Exo and AMI-Exo (Figure 1D). Taken together, these results confirm that exosomes were the main components of the purified microvesicles.
 |
Table 1 The Baseline Characteristics of AMI Patients and Healthy Persons |
Serum-Derived Exosomes Were Internalized Well by Endothelial Cells
To explore whether the exosomes can be internalized by endothelial cells, we incubated PKH67-stained exosomes with HUVECs for 24 h; > 90% of HUVECs were found to be positive for PKH67 fluorescence, with no difference between the internalization of AMI-Exo and Con-Exo (Figure 1E). Confocal imaging confirmed that the exosomes were effectively internalized by the HUVECs (Figure 1F). Thus, the isolated exosomes could be internalized by endothelial cells.
AMI-Exos Promote Angiogenesis in vitro and ex vivo
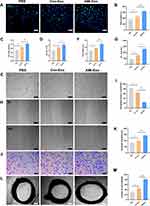
To explore the functions of the exosomes acting on endothelial cells, HUVECs were co-cultured with exosomes isolated from both (control and AMI) groups. AMI-Exo promoted HUVEC proliferation to a greater extent than Con-Exo after treatment, as revealed by EdU and CCK8 assays (Figure 2A-C) and confirmed by manual cell counting (Figure 2D). Vascular formation effect was evaluated using Matrigel assays, which showed that AMI-Exo augment the total tube length and number of nodes compared to Con-Exo, indicating that they possess higher pro-angiogenic abilities (Figure 2E-G). To assess the migration of cells after exosome treatment, we also performed Transwell and scratch wound assays. AMI-Exo exerted greater effects than Con-Exo on both the migration area in the wound assay (Figure 2H and I) and the migrated cell number in the Transwell assay (Figure 2J and K). To further investigate the pro-angiogenic properties of the AMI exosomes, we performed aortic ring assays, which revealed that conditioned media contained AMI-Exo markedly enhanced microvessels sprouting from aortic rings (Figure 2L and M). Overall, results of our in vitro and ex vivo assays showed that both Con-Exo and AMI-Exo could promote angiogenesis compared with PBS group. However, AMI-Exo exerted greater effects than Con-Exo in angiogenesis-related procedures, including proliferation, tube formation, migration, and microvessel sprouting, suggesting that exosomes from the peripheral serum of patients with AMI may contain certain angiogenic signals than those from healthy individuals.
miR-126-3p Expression is Up-Regulated in AMI-Exo
Recent studies have demonstrated that exosomes exert crucial effects in different cell types via miRNA transport. To investigate the mechanisms underlying the angiogenic roles of the exosomes, we chose several angiogenesis-related miRNAs (miR-143, miR-17-5p, miR-21-5p, miR-939-5p, and miR-126-3p),5,12–15 and validated their differential expression in the serum exosomes of AMI patients and control individuals using qRT-PCR. miR-126-3p expression differed significantly between the two groups, indicating that circulating AMI-Exos may load miR-126-3p (Figure 3A-E). Next, HUVECs were incubated with exosomes and RNA polymerase II inhibitor. The results showed that there was no significant difference in the miR-126-3p expression between the two groups (Figure 3F), suggesting that AMI-Exo did not induce endogenous miR-126-3p transcription in HUVECs. The effect induced by miR-126-3p, thus was attributed to exosome transport.
miR-126-3p from AMI-Exo Promotes Angiogenesis in vitro and ex vivo
To verify that miR-126-3p is essential for the exosomal regulation of angiogenesis in vitro and ex vivo, we co-cultured HUVECs and aortic rings with PBS, Con-Exo + mimic NC, Con-Exo + mimic miR-126-3p, AMI-Exo + inhibitor NC, and AMI-Exo + inhibitor miR-126-3p. Con-Exo + mimic miR-126-3p and AMI-Exo + inhibitor NC promoted endothelial cell proliferation better than Con-Exo + mimic NC and AMI-Exo + inhibitor, respectively, as indicated by CCK8 and Edu analyses (Figure 4A-D). In addition, Matrigel assays to assess vascular formation ability revealed that Con-Exo transfected with the miR-126-3p increased the number of nodes and total tube length, whereas AMI-Exo transfected with the inhibitor miR-126-3p decreased AMI-Exo induced vascular formation, suggesting that exosomal miR-126-3p exerts pro-angiogenic effects post AMI (Figure 4E-G). We also performed Transwell and scratch wound assays to evaluate the migration of cells considering the four types of exosomes. Notably, exosomal miR-126-3p transfection promoted cell migration in wound assay (Figure 4H and I) and increased the number of migrated cells in Transwell assay (Figure 4J and K). While, exosomal inhibitor miR-126-3p transfection abolished the effects of AMI-Exo promoted HUVEC migration (Figure 4H-K). Furthermore, conditioned media contained Con-Exo + mimic miR-126-3p or AMI-Exo + inhibitor NC markedly enhanced microvessel sprouting from aortic rings (Figure 4L and M). Together, these findings indicate that miR-126-3p in AMI-Exos plays a key role in angiogenesis.
miR-126-3p from AMI-Exo Promotes Angiogenesis in Mouse Hind Limb Ischemia
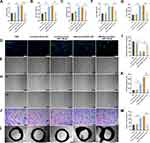
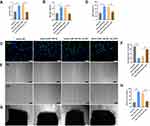
To further investigate the angiogenic effects of miR-126-3p from AMI-Exo and Con-Exo in vivo, we generated a mouse model of unilateral hind limb ischemia that was injected intramuscularly with PBS or exosomes on day 1 after surgery. As per a previous study,5 exosomes were diluted in PBS. Then, we injected 200μL different concentration AMI-Exo, as 5×102 μg/mL, 1×103 μg/mL, 2×103 μg/mL, or PBS on day 1 after surgery. We found that the exosomal concentration of approximately 1×103 μg/mL in 200 μL PBS was the optimal amount (Figure S1). Thus, this amount was chosen for the following experiments. qRT-PCR assays revealed a high miR-126-3p expression in the muscles treated with Con-Exo + mimic miR-126-3p and AMI-Exo + inhibitor NC (Figure 5A). No significant differences in blood flow recovery were observed between the four groups 3 and 7 days post-treatment; however, the Con-Exo + mimic miR-126-3p and AMI-Exo + inhibitor NC groups showed significantly higher blood perfusion 14 days post-injection than the Con-Exo + mimic NC and AMI-Exo + inhibitor miR-126-3p groups, respectively (Figure 5B and C). The gastrocnemius muscles were harvested, embedded, sectioned, and subjected to H&E staining (Figure 5D) and immunofluorescence (Figure 5E). Angiogenesis after ischemia was detected using the vascular marker CD31 and α-SMA. The groups treated with Con-Exo + mimic miR-126-3p and AMI-Exo + inhibitor NC showed a significantly higher capillary density than those treated with Con-Exo + mimic NC and AMI-Exo + inhibitor miR-126-3p (Figure 5F), respectively, indicating that exosomal miR-126-3p may play a key role in microvascular construction. Overall, these analyses of the mouse hind limb ischemia model showed that miR-126-3p in peripheral serum-derived exosomes promotes post-ischemic recovery.
miR-126-3p from AMI-Exo Promotes Angiogenesis in Mouse AMI Model
To further investigate the angiogenic effects of exosomal miR-126-3p in mouse AMI model, we intramyocardially injected PBS or exosomes into the left ventricular wall (border zone) immediately after the left anterior descending ligation. The cardiac tissue was collected 14 days after the myocardial infarction. The qRT-PCR assays revealed that the miR-126-3p-enriched exosomes could effectively deliver miR-126-3p to the myocardium (Figure 6A). The immunohistochemical assay was performed to assess angiogenesis after ischemia by detecting the number of vessels. Con-Exo + mimic miR-126-3p group showed a significantly higher capillary density than Con-Exo + mimic NC group. The AMI-Exo + inhibitor miR-126-3p group showed a significantly lower capillary density than AMI-Exo + inhibitor NC group (Figure 6B–D). Collectively, these analyses of the mouse AMI model showed that miR-126-3p in peripheral serum-derived exosomes promotes post-ischemic endothelial cell angiogenesis.
miR-126-3p Promotes Endothelial Cell Angiogenesis by Targeting TSC1 and Activates the mTORC1/HIF-1α Signaling Pathway
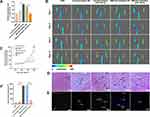
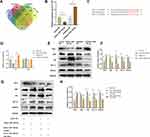
Mutations in TSC1 or TSC2 have been reported to lead to benign malformations or tumors, such as lymphangioleiomyomatosis and angiomyolipoma, in several different organs.16 Therefore, TSC1 may play important roles in angiogenesis. The starBase, miRWalk, miRcode, and miRDB databases predicted that TSC1 is a target gene of miR-126-3p (Figure 7A). The qRT-PCR analyses validated that miR-126-3p overexpression greatly suppresses TSC1 expression in HUVECs, with miR-126-3p inhibition exerting the opposite effects (Figure 7B).
To investigate the binding between miR-126-3p and the 3ʹ UTR of TSC1, we performed luciferase assays in which the TSC1 wild-type 3ʹ UTR or mutant 3ʹ UTRs were cloned into a reporter plasmid and their response to miR-126-3p was assessed in HUVECs (Figure 7C). The mimic miR-126-3p markedly decreased luciferase activity, whereas the inhibitor miR-126-3p increased luciferase activity. Moreover, the mutant 3ʹ UTRs of TSC1 did not alter the response of luciferase activity to the mimic miR-126-3p or inhibitor (Figure 7D). Therefore, these results confirm that TSC1 is a direct target of miR-126-3p.
Further Western blot analyses suggested that miR-126-3p overexpression reduces TSC1 expression, with miR-126-3p inhibition boosting TSC1 expression. TSC1 is known to target the mTORC1/HIF-1α pathway.17 Hence, our next step was to confirm the effects of changes in miR-126-3p expression on the TSC1/mTORC1/HIF-1α pathway. HUVECs treated with mimic of miR-126-3p had improved levels of phosphorylated S6 (pS6) and S6, which are markers of mTORC1 activation, compared to those treated with the control. Conversely, pS6 and S6 levels decreased when the cells were treated with the inhibitor of miR-126-3p. mTORC1 signaling was highly activated in HUVECs along with increase in HIF-1α and VEGFA levels (Figure 7E and F), consistent with previous reports that mTORC1 signaling regulates HIF-1α transcription.18 Subsequently, we constructed a TSC1 overexpression plasmid and simultaneously transfected with miR-126-3p. We found that the miR-126-3p induced mTORC1 activation and increased the expression of HIF-1α and VEGFA, which were abolished by TSC1 overexpression (Figure 7G and H).
To further investigate whether miR-126-3p could modulate angiogenesis by targeting TSC-1, we utilized validation experiment, wherein, miR-126-3p and TSC1 were overexpressed simultaneously to observe the phenotype of angiogenesis. The overexpression of TSC1 abolished miR-126-3p-promoted endothelial cell proliferation effect, as indicated by CCK8 and Edu analyses (Figure 8A–D). In scratch wound assays, the overexpression of TSC1 could reduce the effect of miR-126-3p-promoted cell migration area in wound assay (Figure 8E and F). Furthermore, in aortic ring assay, the overexpression of TSC1 could blocked miR-126-3p induced microvessel sprouting (Figure 8G and H). Collectively, these findings indicate that miR-126-3p promotes endothelial angiogenesis by targeting TSC1.
Discussion
Exosomes carry many functional messages which play key roles in cell-to-cell interactions during myocardial ischemia. MiRNAs are small noncoding RNAs with 22–23 nucleotides that could post-transcriptionally restrain gene expression by targeting the 3ʹ UTR of mRNAs. Notably, exosomal miRNAs have emerged as key mediators in diverse biological and pathological processes, including the promotion of angiogenesis and improvement of cardiac function.18,19 Indeed, recent studies have verified that exosomes containing miRNAs can be delivered into other cells to post-transcriptionally regulate gene expression.20 Exosomes from the coronary artery serum of patients with myocardial ischemia or those with myocardial infarction have been shown to promote angiogenesis through the miR-939-iNOS-NO pathway5 and through the IGF-IR/NO signaling pathway.21 However, exosomes secreted by cells may undergo diverse changes and exert different effects under different pathophysiological conditions,22 and the effects of exosomes derived from the peripheral serum of patients with early stage AMI on angiogenesis remain unclear.
Zhang and Zhang23 demonstrated that mouse cardiomyocytes could release specific exosomes into the bloodstream under ischemic conditions. Therefore, in this study we obtained exosomes from human peripheral serum, which were subsequently internalized by endothelial cells. We found that AMI-Exo enhanced HUVEC proliferation, tube formation, migration, and enhanced microvessel sprouting from aortic rings. It was recently discovered that exosomes contain numerous miRNAs, including miR-143 and miR-126, that participate in various phases of angiogenesis.5,15,21 In particular, miR-126 is expressed at high levels in vascular endothelial cells and is known as a master regulator for vascular integrity and physiological angiogenesis.24,25 Xue et al26 reported that the expression levels of plasma miR-17-5p, miR-126, and miR-145-3p were significantly augmented in patients with AMI within 4 h of the onset of chest pain. Moreover, Ling et al27 reported that serum exosomal miR-126 and miR-21 levels were higher in unstable angina (UA) and AMI patients (within 2 h of AMI) than healthy controls. Moreover, circulating exosomal miR-126 expression levels correlated positively with the severity of coronary artery stenosis defined by Gensini score. Therefore, in this current study, we validated the expression of some angiogenesis-related miRNAs, including miR-143, miR-17-5p, miR-21-5p, miR-939-5p, and miR-126-3p between two groups using qRT-PCR. Among them, the miR-126-3p expression levels were significantly upregulated in AMI-Exo. Moreover, gain-of-function analysis in HUVECs revealed that miR-126-3p significantly promoted cell proliferation, tube formation, migration, and microvessel sprouting. miR-126-3p also improved angiogenesis to a greater extent in AMI-Exo-treated cells than Con-Exo-treated cells in mouse limb ischemia and AMI models.
Jakob et al28 previously revealed that exosomal miR-126 from the endothelial cells of patients with chronic heart failure could affect angiogenesis by regulating SPRED1/VEFG signaling. In neonates, TSC1 loss has been shown to increase retinal angiogenesis and lead to endothelial proliferative impairment and generation of vascular tumors in mice by activating mammalian target of rapamycin (mTOR) complex 1 (mTORC1) and increasing HIF-1α and VEGFA expression.17 Although the post-transcriptional inhibition of TSC1 by miR-126-3p has been reported in granulosa cells,29 the source and delivery route of miR-126-3p during the early stages of AMI remain unknown. Since bioinformatics analysis and a previous study29 have hinted that TSC1 is a promising target gene of miR-126-3p, we used luciferase reporter assays to demonstrate that miR-126-3p directly targets the 3ʹ UTR of TSC1 and represses TSC1 expression.
TSC1 is a subtype of the TSC gene that can form a protein complex with TSC2.30 The mutations in TSC1 and TSC2 have been shown to give rise to the lymphangioleiomyomatosis.31 The complex formed by the TSC1 and TSC2 gene products is a pivotal negative regulator of mTORC1 and cell growth.32 mTOR is a serine/threonine kinase which regulates a number of cellular processes,33 and any abnormality in mTORC1 signaling can result in many diseases including vascular malformations and even tumors.34,35 Studies have shown the key role of VEGF in tumor development regulated by HIF-1α, which is present downstream of mTORC1,17 and that HIF-1α is an essential mediator of VEGF-induced angiogenesis.36,37 Consistently, we observed that miR-126-3p targeted TSC1, which activated the mTORC1 signaling pathway, increased pS6 and S6 expression, and increased HIF-1α and VEGFA expression. Interestingly, HIF-1α upregulation has been shown to affect the regulated miR-126 expression upstream that can promote angiogenesis and subsequently improve heart function in myocardial infarction.38 Therefore, there may be a previously undescribed mechanism underlying the promotion of endothelial angiogenesis that involves a positive feedback loop of miR-126-3p and HIF-1α, and this should be verified in future studies.
Previously, Nie et al39 reported that circulating miR-126-3p levels were significantly higher in the plasma from sufficient coronary collateral circulation in patients with severely narrowed coronary arteries. Unlike the targeting function of exosomal miRNA, circulating miRNA acts as a rover in the circulatory system that can react with any cell type that is in contact with the blood stream. Therefore, the findings of this study suggest that exosome-mediated miR-126-3p delivery to endothelial cells could better explain the improved coronary collateral circulation in patients with higher miR-126-3p levels.
However, this study also has some limitations. First, we focused on the expression of only few miRNAs in exosomes, which might have resulted in the neglect of other functional miRNAs present in patients with AMI. Second, because of the complicated contents of exosomes, the miR-126-3p/TSC1/mTORC1/HIF-1α pathway may be just one of multiple pathways that regulate angiogenesis.
Conclusion
In summary, our results revealed that miR-126-3p in exosomes derived from the peripheral serum of AMI patients can promote angiogenesis in endothelial cells by targeting TSC1 and activating the mTORC1/ HIF-1α axis. Together, the findings of this study provide a novel understanding of the function of exosomal miRNAs during the early stages of AMI onset.
Ethics Approval and Informed Consent
The research protocol was approved by the Second Affiliated Hospital of Harbin Medical University Ethics Committee (KY2021-147, SYDW2020-030).
We obtained written informed consent from all subjects.
Consent for Publication
We obtained written informed consent from all subjects.
Acknowledgments
We thank the Natural Science Foundation of Inner Mongolia, National Natural Science Foundation of China, and the Fund of Key Laboratory of Myocardial Ischemia, Ministry of Education.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was financially supported by the Natural Science Foundation of Inner Mongolia [2019MS08148], the National Natural Science Foundation of China [82102068], and the Fund of Key Laboratory of Myocardial Ischemia, Ministry of Education [KF202106].
Disclosure
The authors report no conflicts of interest for this work.
References
1. Virani SS, Alonso A, Aparicio HJ, et al. Heart disease and stroke statistics-2021 update: a report from the American heart association. Circulation. 2021;143(8):e254–e743. doi:10.1161/CIR.0000000000000950
2. Zhu W, Sun L, Zhao P, et al. Macrophage migration inhibitory factor facilitates the therapeutic efficacy of mesenchymal stem cells derived exosomes in acute myocardial infarction through upregulating miR-133a-3p. J Nanobiotechnol. 2021;19(1):61. doi:10.1186/s12951-021-00808-5
3. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi:10.1080/20013078.2018.1535750
4. Vicencio JM, Yellon DM, Sivaraman V, et al. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol. 2015;65(15):1525–1536. doi:10.1016/j.jacc.2015.02.026
5. Li H, Liao Y, Gao L, et al. Coronary serum exosomes derived from patients with myocardial ischemia regulate angiogenesis through the miR-939-mediated nitric oxide signaling pathway. Theranostics. 2018;8(8):2079–2093. doi:10.7150/thno.21895
6. Pan Q, Wang Y, Lan Q, et al. Exosomes derived from mesenchymal stem cells ameliorate hypoxia/reoxygenation-injured ECs via transferring microRNA-126. Stem Cells Int. 2019:2831756. doi:10.1155/2019/2831756
7. Cao C, Wang B, Tang J, et al. Circulating exosomes repair endothelial cell damage by delivering miR-193a-5p. J Cell Mol Med. 2021;25(4):2176–2189. doi:10.1111/jcmm.16202
8. Thygesen K, Alpert JS, Jaffe AS, et al. Third universal definition of myocardial infarction. J Am Coll Cardiol. 2012;60(16):1581–1598. doi:10.1016/j.jacc.2012.08.001
9. Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542(7642):450–455. doi:10.1038/nature21365
10. Zippel N, Ding Y, Fleming I. A modified aortic ring assay to assess angiogenic potential in vitro. Methods Mol Biol. 2016;1430:205–219. doi:10.1007/978-1-4939-3628-1_14
11. Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi:10.1038/nature14581
12. Tian Y, Li X, Bai C, Yang Z, Zhang L, Luo J. MiR-17-5p promotes the endothelialization of endothelial progenitor cells to facilitate the vascular repair of aneurysm by regulating PTEN-mediated PI3K/AKT/VEGFA pathway. Cell Cycle. 2020;19(24):3608–3621. doi:10.1080/15384101.2020.1857958
13. Jin YP, Hu YP, Wu XS, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018;9(2):182. doi:10.1038/s41419-017-0258-2
14. He Q, Ye A, Ye W, et al. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021;12(6):576. doi:10.1038/s41419-021-03803-8
15. Mathiyalagan P, Liang Y, Kim D, et al. Angiogenic mechanisms of human CD34(+) stem cell exosomes in the repair of ischemic hindlimb. Circ Res. 2017;120(9):1466–1476. doi:10.1161/CIRCRESAHA.116.310557
16. Peter B, Crino KLN, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356.
17. Sun S, Chen S, Liu F, et al. Constitutive activation of mTORC1 in endothelial cells leads to the development and progression of lymphangiosarcoma through VEGF autocrine signaling. Cancer Cell. 2015;28(6):758–772. doi:10.1016/j.ccell.2015.10.004
18. Kenta Hirai DO, Fukushima Y, Kondo M, et al. Cardiosphere-derived exosomal microRNAs for myocardial repair in pediatric dilated cardiomyopathy. Sci Transl Med. 2020;12(573):8764. doi:10.1126/scitranslmed.abb3336
19. Yang H, Zhang H, Ge S, et al. Exosome-derived miR-130a activates angiogenesis in gastric cancer by targeting C-MYB in vascular endothelial cells. Mol Ther. 2018;26(10):2466–2475. doi:10.1016/j.ymthe.2018.07.023
20. Meng W, Hao Y, He C, Li L, Zhu G. Exosome-orchestrated hypoxic tumor microenvironment. Mol Cancer. 2019;18(1):57. doi:10.1186/s12943-019-0982-6
21. Geng T, Song Z, Xing J, Wang B, Dai S. Exosome derived from coronary serum of patients with myocardial infarction promotes angiogenesis through the miRNA-143/IGF-IR pathway. Int J Nanomedicine. 2020;15:2647–2658. doi:10.2147/ijn.S242908
22. McBride JD, Rodriguez-Menocal L, Badiavas EV. Extracellular vesicles as biomarkers and therapeutics in dermatology: a focus on exosomes. J Invest Dermatol. 2017;137(8):1622–1629. doi:10.1016/j.jid.2017.04.021
23. Zhang J, Zhang X. Ischaemic preconditioning-induced serum exosomes protect against myocardial ischaemia/reperfusion injury in rats by activating the PI3K/AKT signalling pathway. Cell Biochem Funct. 2021;39(2):287–295. doi:10.1002/cbf.3578
24. Wang Y, Sun J, Kahaleh B. Epigenetic down-regulation of microRNA-126 in scleroderma endothelial cells is associated with impaired responses to VEGF and defective angiogenesis. J Cell Mol Med. 2021;25(14):7078–7088. doi:10.1111/jcmm.16727
25. Zhou Q, Anderson C, Hanus J, et al. Strand and cell type-specific function of microRNA-126 in angiogenesis. Mol Ther. 2016;24(10):1823–1835. doi:10.1038/mt.2016.108
26. Xue S, Liu D, Zhu W, et al. Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p are novel biomarkers for diagnosis of acute myocardial infarction. Front Physiol. 2019;10:123. doi:10.3389/fphys.2019.00123
27. Ling H, Guo Z, Shi Y, Zhang L, Song C. Serum exosomal MicroRNA-21, MicroRNA-126, and PTEN are novel biomarkers for diagnosis of acute coronary syndrome. Front Physiol. 2020;11:654. doi:10.3389/fphys.2020.00654
28. Jakob P, Doerries C, Briand S, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012;126(25):2962–2975. doi:10.1161/CIRCULATIONAHA.112.093906
29. Yuan X, Deng X, Zhou X, et al. MiR-126-3p promotes the cell proliferation and inhibits the cell apoptosis by targeting TSC1 in the porcine granulosa cells. Vitro Cell Dev Biol Anim. 2018;54(10):715–724. doi:10.1007/s11626-018-0292-0
30. Marjon van Slegtenhorst MN, Nagelkerken B, Cheadle J, Snell R. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7(6):1053–1057. doi:10.1093/hmg/7.6.1053
31. Huang J, Wu S, Wu CL, Manning BD. Signaling events downstream of mammalian target of rapamycin complex 2 are attenuated in cells and tumors deficient for the tuberous sclerosis complex tumor suppressors. Cancer Res. 2009;69(15):6107–6114. doi:10.1158/0008-5472.CAN-09-0975
32. Nisreen El-Hashemite VW, Zhang H, Kwiatkowski DJ. Loss of Tsc1 or Tsc2 induces vascular endothelial growth factor production through mammalian target of rapamycin. Cancer Res. 2003;63:5173–5177.
33. Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem. 2010;285(19):14071–14077. doi:10.1074/jbc.R109.094003
34. Du W, Gerald D, Perruzzi CA, et al. Vascular tumors have increased p70 S6-kinase activation and are inhibited by topical rapamycin. Lab Invest. 2013;93(10):1115–1127. doi:10.1038/labinvest.2013.98
35. Shirazi F, Cohen C, Fried L, Arbiser JL. Mammalian target of rapamycin (mTOR) is activated in cutaneous vascular malformations in vivo. Lymphat Res Biol. 2007;5(4):233–236. doi:10.1089/lrb.2007.1012
36. Rattner A, Williams J, Nathans J. Roles of HIFs and VEGF in angiogenesis in the retina and brain. J Clin Invest. 2019;129(9):3807–3820. doi:10.1172/JCI126655
37. Tian X, Liu Y, Wang Z, Wu S. miR-144 delivered by nasopharyngeal carcinoma-derived EVs stimulates angiogenesis through the FBXW7/HIF-1α/VEGF-A axis. Mol Therapy. 2021;24:1000–1011. doi:10.1016/j.omtn.2021.03.016
38. Song W, Liang Q, Cai M, Tian Z. HIF-1alpha-induced up-regulation of microRNA-126 contributes to the effectiveness of exercise training on myocardial angiogenesis in myocardial infarction rats. J Cell Mol Med. 2020;24(22):12970–12979. doi:10.1111/jcmm.15892
39. Xiaomin Nie LS, Zhou Y, Zhao Y. Association between plasma levels of microRNA-126 and coronary collaterals in patients with coronary artery disease. Chin J Cardiol. 2014;42(7):561–565.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.