Back to Journals » International Journal of Nanomedicine » Volume 18
Periodontal Ligament Stem Cell Exosomes Key to Regulate Periodontal Regeneration by miR-31-5p in Mice Model
Authors Lu J, Yu N, Liu Q, Xie Y, Zhen L
Received 24 May 2023
Accepted for publication 6 September 2023
Published 18 September 2023 Volume 2023:18 Pages 5327—5342
DOI https://doi.org/10.2147/IJN.S409664
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Jiuqing Lu,1,2,* Nijia Yu,1,* Qian Liu,2,* Yajia Xie,1 Lei Zhen2
1Oral Biomedical Engineering Laboratory, Shanghai Stomatological Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Stomatology, Tongji Hospital, School of Medicine, Tongji University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lei Zhen, Department of Stomatology, Tongji Hospital, School of Medicine, Tongji University, Shanghai, People’s Republic of China, Tel +86 1 582 108 9038, Email [email protected]
Introduction: Periodontitis is a chronic inflammatory disease that causes alveolar bone loss. Diabetes is one of the most important factors contributing to periodontitis. Exosomes derived from mesenchymal stem cells (MSCs-Exo) have been reported to promote bone regeneration. This study aimed to examine the function and mechanism of exosomes derived from periodontal ligament stem cells (PDLSCs-Exo) in regulating periodontal regeneration in diabetic periodontitis.
Methods: Exosomes derived from normal-glucose-cultured PDLSCs (NG-PDLSCs-Exo) and high-glucose-preconditioned PDLSCs (HG-PDLSCs-Exo) were used. Their effects on RAW264.7 cells were investigated by TRAP staining and quantitative real time-polymerase chain reaction (qRT-PCR). The role of exosomal miR-31-5p in osteoclast differentiation was tested using qRT-PCR, double luciferase analysis, and Western blotting. We investigated the effects of these two types of PDLSCs-Exo on alveolar bone loss in vivo in mice with experimental periodontitis.
Results: PDLSCs-Exo were transferred to RAW264.7, and HG-PDLSCs-Exo inhibited osteoclast formation to a lesser extent than NG-PDLSCs-Exo. Further studies revealed the effect of PDLSCs-Exo on osteoclastogenesis via the miR-31-5p/eNOS signaling pathway. In mice with experimental periodontitis, PDLSCs-Exo reduced alveolar bone destruction and decreased the number of osteoclasts on the alveolar bone surface.
Conclusion: Our results suggest that exosomal miR-31-5p derived from PDLSCs regulates alveolar bone regeneration by targeting eNOS.
Keywords: diabetes, periodontitis, macrophage, osteoclast differentiation
Introduction
Chronic periodontitis (CP) is a chronic inflammatory disease that leads to the breakdown of the supporting tissue and alveolar bone loss.1 This condition is closely related to various systemic diseases, including diabetes.2 Large pidemiological studies showed that diabetes increases susceptibility to periodontitis.3,4 The risk of alveolar bone loss in patients with diabetic periodontitis is significantly elevated compared to non-diabetic periodontitis; however, the underlying mechanism remains elusive.5,6
Periodontal ligament stem cells (PDLSCs) are a type of mesenchymal stem cells existing in periodontal tissues that have the characteristics of self-renewal, multi-differentiation, immune regulation, and rapid proliferation.7,8 It has been reported that periodontal ligament stem cells are important cell sources for alveolar bone regeneration.9 Chen et al conducted a randomized clinical trial, which demonstrated a significant increase in alveolar bone height across all experimental groups when using autologous periodontal stem cells for the treatment of alveolar bone defects in periodontitis.10 Ma et al reported that compared with hydrogels alone, the incorporation of PDLSCs into hydrogels enhanced the new bone formation of alveolar bone defects in rats.11 Another study revealed the osteogenic and lipid properties of periodontal ligament stem cells, while in vivo transplantation models demonstrated their capacity to facilitate periodontal ligament fiber regeneration accompanied by the formation of new bone-like tissue.12
Exosomes (Exos) are a class of extracellular vesicles with a diameter of 50–150 nm that are rich in proteins, lipids, RNA, miRNA, mRNA, etc., and act as carriers for delivering signaling molecules to mediate cell-to-cell communication.13,14 Similar to the regenerative potential of stem cells, exosomes possess the ability to promote bone regeneration.15 Lei et al demonstrated that exosomes derived from human periodontal ligament stem cells (hPDLSCs-Exo) effectively enhanced bone regeneration in rats with alveolar bone defects and mitigated inflammatory bone responses.16 Yang et al revealed that, in comparison to the control group and the hydrogel group, rats treated with PDLSCs-Exo hydrogel exhibited significantly enhanced new bone formation in alveolar bone defects.17
Moreover, exosomal miRNAs are an important mechanism of exosome-mediated bone remodeling.18 MSCs-Exo can promote osteoblast differentiation and proliferation in vitro through miR-1260a and promote bone formation in SD rats with skull defects in vivo.19 Exosomes derived from human adipose-derived stem cells (hASCs-Exo) overexpressing miR-375 improved osteogenic differentiation of human bone marrow mesenchymal stromal cells, and further in vivo analysis showed that exosomal miR-375, combined with hydrogel enhanced bone regeneration in a rat model of calvarial defects.20,21 Exosomes derived from PGE2-induced PDLSCs inhibit the osteogenic differentiation of human periodontal ligament stem cells by activating the miR-34c-5p/SATB2/ERK pathway.22 These findings provide preliminary evidence for the pivotal role of exosomal miRNAs in bone regeneration. However, the precise function of exosomal miRNAs derived from PDLSCs in alveolar bone regeneration remains unclear.
The miRNA chip sequencing analysis conducted in our previous study demonstrated that miR-31-5p exhibited the highest level of upregulation in periodontal ligament stem cells cultured under high-glucose conditions (HG-PDLSCs), and the upregulated miR-31-5p expression in PDLSCs might be related to the weakened osteogenic differentiation ability.23 In this study, we aimed to compared the effects of NG-PDLSCs-Exo and HG-PDLSCs-Exo on alveolar bone regeneration and related miRNA mechanism. The regenerative potential of HG-PDLSCs-Exo in periodontal tissue was comparatively weaker than that of NG-PDLSCs-Exo, which is due to the relatively diminished capacity of HG-PDLSCs-Exo to suppress osteoclastogenesis. Subsequent investigations revealed that PDLSCs-Exo effectively targeted eNOS by delivering miR-31-5p, thereby influencing osteoclastogenesis in a high-glucose microenvironment.
Materials and Methods
The Cell Culture Studies
This study was approved by the Ethics Committee of the Shanghai Stomatological Hospital, Fudan University. Ten healthy premolars requiring extraction due to orthodontics were collected from patients aged 15–30 years at the Department of Oral and Maxillofacial Surgery of Shanghai Stomatological Hospital, Fudan University. Informed consent was obtained from all patients or their guardians who signed informed consent forms, and the periodontal ligament in the 1/3 area of the root was gently scraped with a 11# surgical blade and cut into 1 mm³ size tissue. The tissue was digested with 2 mL of 3 mg/mL type I collagenase (Sigma, Louis, USA) for 60 min and incubated in Dulbecco modified Eagle medium (DMEM) (Gibco, California, USA) containing 10% FBS (Gibco, California, USA).
Cells were purified using a limited-dilution technique. Cells in the logarithmic growth phase were treated with pancreatic enzymes (Gibco, California, USA), and the cell suspension was diluted to a concentration of 5 cells/mL. Subsequently, an inoculum volume of 100 μL per well was added to 96-well plates. Single-cell wells were observed under a microscope after 24 h. After 1–2 weeks of cultivation, the clones produced from single cell were collected and pooled. PDLSCs at passages 2–4 were selected for subsequent experiments. After one passage, cells were harvested for identification. For adipogenic and osteogenic differentiation, passage 2 PDLSCs were cultured in 6-well plates. After reaching 90% confluence, cells were directionally induced for 2-3 weeks using Stem Cell Adipogenic Induction Kit (Cyagen Biosciences, California, USA) and Stem Cell Osteogenic Induction Kit (Cyagen Biosciences, California, USA). After that, cells were fixed for 20 min in 4% paraformaldehyde (Beyotime Biotechnology, Shanghai, China), and then stained with 0.5% Oil Red O (Sigma, Louis, USA) and 0.2% Alizarin Red S (Sigma, Louis, USA) solutions.
Immunofluorescence staining was used to detect the cell surface markers. Passage 2 PDLSCs were cultured in 24-well plates. Cells were immunolabeled for 4 hours with STRO-1 antibody (R&D Systems, Minneapolis, MN, USA) (1:1000) when they reached 80% confluence. Cells were visualized using a fluorescent secondary antibody (Jackson ImmunoResearch, Pennsylvania, USA) (1:1000). Finally, 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology, Shanghai, China) was used to stain the nuclei. For flow cytometry analysis, 5×106 PDLSCs at passage 2 were suspended in 900 μL of PBS and divided into 5 portions. One portion was added to 500 μL of PBS as a blank control, and the other portions were added to 100 μL of rabbit polyclonal anti-CD73, anti-CD90, anti-CD34, and anti-CD45 antibodies (BioLegend, California, USA) (1:500) on ice for 2 h away from light. Finally, the antibody was removed by centrifugation, and cells precipitates were resuspended in 500 µL PBS for detection by flow cytometry (Beckman Coulter, California, USA).
The Isolation and Examination of Exosomes, Using Ultracentrifugation, Western Blot, Transmission Electron Microscopy, and Visual Nanoparticle Analyzer
Passage 3 PDLSCs were cultured in 10-cm dishes. After reaching 90% confluence, the cells were divided into 2 groups: PDLSCs were cultured in a medium with 5.5 mM glucose and 2% exosome-free FBS (SBI, USA), referred to as NG-PDLSCs; and PDLSCs were cultured in a medium with 33 mM glucose and 2% exosome-free FBS, referred to as HG-PDLSCs. After 48 h of culture, the supernatants of NG-PDLSCs and HG-PDLSCs were ultra centrifuged (Beckman Coulter, California, USA) under 4 °C at 300 × g for 10 min, 2000 × g for 10 min, 10,000 × g for 30 min, and 100,000 × g for 70 min. The precipitates were resuspended in 200 μL precooled PBS and analyzed by the BCA Protein Assay (Beyotime Biotechnology, Shanghai, China). The morphology of the exosomes was observed through transmission electron microscopy (TEM) (Hitachi, Tokyo, Japan), and the diameter of exosomes was measured using visual nanoparticle analyzer (Beckman Coulter, California, USA). In addition, exosomal surface markers, including rabbit polyclonal anti-CD9 and rabbit polyclonal anti-CD63 antibodies (Beyotime Biotechnology, Shanghai, China), were detected by Western blotting.
The Label and Uptake of Exosomes, Using Immunostaining and Immunofluorescence
NG-PDLSCs-Exo and HG-PDLSCs-Exo were labeled using a PKH67 Kit (Sigma, Louis, USA). The labeled exosomes were re-isolated and co-cultured with RAW264.7 cells purchased from National Collection of Authenticated Cell Cultures (Shanghai, China) at a concentration of 5 μg/mL for 24 hours. A Red fluorescent probe from a microfilament (Beyotime Biotechnology, Shanghai, China) was used to label the cell membrane, and DAPI was used to stain the nuclei. Finally, the uptake of exosomes was observed under an inverted fluorescence microscope (Leica, Wetzlar, Germany).
The Culture and Treatment of RAW264.7
The RAW264.7 cells were cultured in alpha minimal essential medium (α-MEM) (Gibco, California, USA) with 10% FBS. At 90% confluence, the cells were treated with 50 ng/mL macrophage colony-stimulating factor (M-CSF) (R&D Systems, Minneapolis, MN, USA) for 3 days and 50 ng/mL receptor activator of nuclear factor-κB ligand (RANKL) (R&D Systems, Minneapolis, MN, USA) for 4 days. They were also co-cultured with exosomes at a concentration of 15 μg/mL. The experimental groups included the control, induce, induce+ NG-PDLSCs-Exo, and induce + HG-PDLSCs-Exo.The area of osteoclast formation was assessed using a TRAP staining kit (Wako, Tokyo, Japan), and the genes related to osteoclastogenesis were examined using quantitative real time-polymerase chain reaction (qRT-PCR) (Roche, Basel, Switzerland).
Animal Experiments
All animal experiments were approved by the Ethics Committee of the Shanghai Stomatological Hospital, Fudan University. All animal experiments are conducted in accordance with the guidelines of local regulatory agencies and comply with the “Regulations on the Management of Experimental Animals” issued by the Ministry of Science and Technology of the People’s Republic of China. Twenty-four C57BL/6 male mice, aged 6–8 weeks, were purchased from Shanghai Jiesjie Laboratory Animal Co. Ltd. All mice were raised in a clean-level animal house at the Fudan University. Then, mice were treated with 50 mg/kg pentobarbital sodium for anesthesia prior to induction of the experimental periodontitis model. The maxillary first molars were ligated using a 6–0 suture (Jinhuan, Shanghai, China), and the suture was pressed into the gingival sulcus. On the same day as modeling, exosomes were injected into the periodontal membrane of the maxillary first molars. The exosomes were administered via periodontal injection at six sites on the maxillary first molars: mesiobuccal, midbuccal, distobuccal, mesiopalatal, midpalatal, and distopalatal. The dosage regimen consisted of a weekly administration of 300 μg for 4 weeks. The experiment was divided into 4 groups: blank control (n = 6), chronic periodontitis (CP) (n = 6), chronic periodontitis + NG-PDLSCs-Exo (CP+NG-PDLSCs-Exo) (n = 6), and chronic periodontitis + HG-PDLSCs-Exo (CP+HG-PDLSCs-Exo) (n = 6). Finally, the mice were euthanized by over-anesthetization after four weeks of exosomal treatment.
Methylene Blue Staining
The soft tissues surrounding the maxillary samples were removed. Resorption of the alveolar bone was observed under a stereomicroscope (Leica, Wetzlar, Germany) after staining with a 1% methylene blue solution (Sigma, Louis, USA) for 1 min, followed by washing with PBS for 5 min.
Micro-CT Analysis
Four ipsilateral maxillary samples were randomly selected from each group for analysis of alveolar bone resorption using micro-CT (Bruker, Kontich, Belgium). The level of bone loss in the maxillary first molars was quantitatively assessed by measuring cementoenamel junctions to the alveolar bone crest (CEJ-ABC) at six sites. CTan software was used to analyze the bone mineral density in the distal alveolar bone of the maxillary first molar.
Histological Analysis
The maxilla was fixed, dehydrated, embedded, and sectioned at a thickness of 5 μm. Subsequently, the sections were stained with a hematoxylin and eosin (H&E) staining kit (Beyotime Biotechnology, Shanghai, China) and TRAP staining kit (Servicebio, Wuhan, China) for histological analysis.
RNA Interference
PDLSCs were transiently transfected with miR-31-5p nc/mimics/inhibitors (GenePharma, Shanghai, China) using siRNA-Mate (GenePharma, Shanghai, China). Subsequently, exosomes were isolated from the supernatant of transfected PDLSCs, and their effects on osteoclast differentiation of RAW264.7 cells was evaluated through qRT-PCR and TRAP staining.
Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the cells and exosomes with TRIzol reagent (Tiangen, Beijing, China). For mRNA analysis, cDNA was synthesized using the Fastking RT Kit (Tiangen, Beijing, China), followed by qRT-PCR using the SuperReal PreMix Plus Kit (Tiangen, Beijing, China) on a LightCycler®96 instrument (Roche, Basel, Switzerland). For miRNA analysis, cDNA synthesis was performed using MiRcute Enhanced miRNA cDNA First Strand Synthesis Kit (Tiangen, Beijing, China) and qRT-PCR was conducted using MiRcute Enhanced miRNA Fluorescence Quantitative Detection Kit (Tiangen, Beijing, China) on a LightCycler®96 instrument. The mRNA expression levels of the NFATc1, TRAF6, and c-Fos genes were quantified by the 2−ΔΔCt method using β-actin as a reference gene. The miRNA expression levels of miR-31-5p were determined via the 2−ΔΔCt method using U6 as a reference. Table 1 shows the list of primers.
 |
Table 1 List of Specific Primers |
Dual Luciferase Reporter Assay
The eNOS 3’-UTR sequence was amplified by primers designed on GenBank (human eNOS, NM_000603.4), and then ligated into the MCS of pGL3-control plasmid via EcoRI and XbaI restriction sites. The plasmid PGL3-WT-eNOS carrying the wild-type eNOS 3’-UTR was obtained after digestion and subsequent sequence verification. Using overlap PCR, the mutant plasmid PGL3-MT-eNOS was generated by targeted deletion of the miR-31 seed sequence binding site on PGL3-WT-eNOS. The luciferase activity of the dual 1.5 dual luciferase reporter plasmid was measured with a luciferase reporter assay system kit (Thermo Scientific, Massachusetts, USA). Firefly luciferase recognition scores were normalized to Renilla luciferase as an internal reference and statistical significance was determined by comparison to the control group.
Western Blot
The total protein content of the exosomes and cells was extracted with RIPA Lysis Buffer (Beyotime Biotechnology, Shanghai, China) and quantified with the BCA Protein Assay. Proteins were separated by 4–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Thermo Scientific, Massachusetts, USA) with a loading of 20 μg total protein per well.The Proteins were transferred from the gel to a PVDF membrane. The PVDF membranes were washed with blocking buffer for 20 min and incubated for 4 h at room temperature. The primary antibodies used in this study were as follows: rabbit polyclonal anti-CD9 (Beyotime Biotechnology, Shanghai, China) (1:1000), rabbit polyclonal anti-CD63 (Beyotime Biotechnology, Shanghai, China)(1:1000), eNOS (Affinity, Connecticut, USA) (1:1000), and the internal reference GAPDH (Beyotime Biotechnology, Shanghai, China) (1:2000). The corresponding secondary antibodies were then incubated at room temperature for 2 h. After the TBS-T washes, a chemiluminescent reagent was added and then exposed to X-ray film.
Statistical Analysis
Data are presented as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) were used for comparisons between different groups. All calculations were performed using GraphPad Prism software (version 8.0; GraphPad Software, San Diego, CA, USA). Statistical significance was defined as P < 0.05.
Results
Identification of PDLSCs
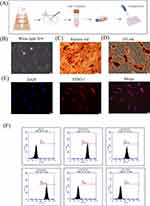
PDLSCs were successfully isolated and cultured using an enzymatic digestion method (Figure 1A). The PDLSCs exhibited a long spindle shape under a microscope (Figure 1B). Alizarin red S staining demonstrated the formation of calcium nodules following osteogenic induction (Figure 1C). Oil Red O staining showed the formation of lipid droplets after adipogenic induction (Figure 1D). The immunofluorescence results showed that the specific antigen STRO-1 was highly expressed in PDLSCs (Figure 1E). Flow cytometry showed that PDLSCs were positive for CD73 and CD90 and negative for CD34 and CD45, which is consistent with the characteristics of mesenchymal stem cells (Figure 1F).
Identification of Exosomes
The exosomes were separated by ultracentrifugation (Figure 2A). Both NG-PDLSCs-Exo and HG-PDLSCs-Exo exhibited a circular or elliptical morphology with a bilayer structure, as observed by electron microscopy (Figure 2B), and their diameters were approximately 50–150 nm (Figure 2C). Specific marker proteins, CD9 and CD63, were expressed on their surfaces and met the criteria for exosome identification (Figure 2D).
NG-PDLSCs-Exo Significantly Inhibited Osteoclast Differentiation in vitro Compared to HG-PDLSCs-Exo
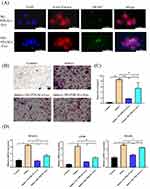
Immunofluorescence analysis showed that NG-PDLSCs-Exo and HG-PDLSCs-Exo were successfully taken up by RAW264.7 cells (Figure 3A). TRAP staining analysis revealed the presence of red multinucleated giant cells after osteoclast induction of RAW264.7 cells, consistent with results reported in previous studies.24,25 In addition, we demonstrated a significant reduction in red multinucleated giant cells formation in the Induce+NG-PDLSCs-Exo group compared to Induce+HG-PDLSCs-Exo group (Figure 3B). Furthermore, the area occupied by the red multinucleated giant cells was significantly smaller in the Induce+NG-PDLSCs-Exo group than in the Induce+HG-PDLSCs-Exo group (Figure 3C). qRT-PCR showed that NG-PDLSCs-Exo downregulated the osteoclast genes NFATc1, TRAF6, and c-Fos, while HG-PDLSCs-Exo downregulated only the osteoclast gene TRAF6 (Figure 3D).
NG-PDLSCs-Exo Significantly Attenuated Alveolar Bone Loss in Mice with Experimental Periodontitis Compared to HG-PDLSCs-Exo
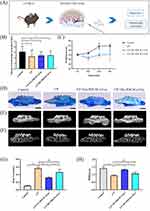
Experimental periodontitis was successfully induced in the mice by ligating the maxillary first molars using a 6–0 suture (Figure 4A). The mice exhibited a decrease in food intake and a small initial decrease in body weight (Figure 4B and C). Methylene blue staining and micro-CT showed significant alveolar bone loss in the CP group, whereas the degree of alveolar bone loss was less in the CP+NG-PDLSCs-Exo group than in the CP+HG-PDLSCs-Exo group (Figure 4D–H). HE and TRAP staining results demonstrated that the CP+NG-PDLSCs-Exo group showed a significant improvement in alveolar bone resorption, and osteoclasts were almost invisible in the alveolar bone, whereas the CP+HG-PDLSCs-Exo group showed no significant improvement in alveolar bone resorption, and a certain number of osteoclasts still infiltrated the alveolar bone (Figure 5).
PDLSCs-Exo Regulated Osteoclast Formation Through the Transfer of miR-31-5p
qRT-PCR showed that HG-PDLSCs highly expressed miR-31-5p (Figure 6A) and HG-PDLSCs-Exo also highly expressed miR-31-5p (Figure 6B). In addition, miR-31-5p was also highly expressed in RAW264.7 cells co-cultured with HG-PDLSCs-Exo (Figure 6C). Cell models with both overexpression and repression of miR-31-5p in PDLSCs were established by cell transfection (Figure 6D), resulting in the corresponding exosomes displaying altered levels of miR-31-5p expression (Figure 6E). In addition, miR-31-5p was highly expressed in RAW264.7 cells co-cultured with miR-31-mimics-Exo (Figure 6F). TRAP staining revealed a significantly higher incidence of red multinucleated giant cells in the Induce+miR-31 mimics-Exo group than in the Induce+miR-31 inhibitors-Exo group (Figure 6G). Moreover, the area occupied by the red multinucleated giant cells was significantly larger in the Induce+miR-31 mimics-Exo group than in the Induce+miR-31 inhibitors-Exo group (Figure 6H). qRT-PCR showed that exosomes overexpressing miR-31-5p upregulated the osteoclast genes NFATc1, TRAF6, and c-Fos, whereas exosomes with repression of miR-31-5p downregulated the osteoclast genes NFATc1, TRAF6, and c-Fos (Figure 6I).
MiR-31-5p Directly Targeted eNOS
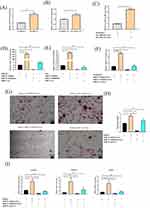
We then examined the mechanism underlying miR-31-5p-mediated promotion of osteoclast function. Online prediction tools such as TargetScan (http://www.targetscan.org/vert70/), miRanda (http://www.microrna.org/), and miRDB (http://mirdb.org/), were used to identify the predicted targets of this miRNA (Figure 7A). Twenty predicted targets were identified. As shown in Figure 7B and C, GO and KEGG enrichment analyses of miR-31-5p showed the involvement of biological processes (insulin resistance, citrate cycle, protein-containing complex disassembly, etc.) and signaling pathways(canonical Wnt signaling pathway, intracellular receptor signaling pathway, regulation of pluripotency of stem cell signaling pathways, etc.) possibly related to osteoclastogenesis. eNOS has been identified as a target gene associated with osteoclastogenesis. We next examined its functional role in miR-31-5p-mediated osteoclastogenesis. We began performing a luciferase reporter assay, which confirmed that luciferase activity in eNOS-WT cells was inhibited by the miR-31-5p mimics (Figure 7D and E). Western blot results also confirmed that eNOS protein expression was significantly reduced in osteoclasts transfected with miR-31-5p mimics compared to the negative control (Figure 7F and G).
Discussion
This study elucidates the role of PDLSC-derived exosomal miRNAs in periodontitis and offers a new strategy to treat alveolar bone regeneration in diabetic periodontitis. In particular, exosomes isolated from PDLSCs in normal glucose had a relatively strong inhibitory effect on osteoclast formation in vitro, whereas exosomes isolated from PDLSCs in high glucose had a weak inhibitory effect on osteoclast formation. Furthermore, the effect of exosomes on osteoclast formation was mediated through the upregulation of miR-31-5p, which is related to eNOS targeting.
Tissue breakdown and alveolar bone loss are characteristic of periodontitis.1 Moreover, the progression of periodontitis may be accelerated in the presence of hyperglycemia.1 It has been reported that stem cell-derived exosomes have demonstrated the potential to enhance periodontal tissue regeneration and mitigate periodontal degeneration.26 Exosomes derived from human exfoliated deciduous stem cells (SHEDs) could enhance osteogenic differentiation of PDLSCs and promote periodontal tissue regeneration.27 Another study found that exosomes secreted by MSCs accelerated the formation of new periodontal tissue in experimental models of periodontitis by promoting AKT and activating ERK signaling.28 Lan et al found that PDLSCs-Exo can promote proliferation, migration and osteogenic differentiation of hfob 1.19 cells, inhibit H2O2-induced apoptosis, and activate PI3K/AKT and MEK/ERK signaling pathways, which may be a promising method for the treatment of osteoblast injury.29 In this study, we isolated, purified, and characterized exosomes from PDLSCs. Furthermore, we demonstrated that exosomes derived from periodontal ligament stem cells regulate periodontal regeneration in mice model.
However, the microenvironment can significantly affect the secretion and biological functions of exosomes.30–32 Exosomes obtained from hypoxia-conditioned SHEDs enhanced angiogenesis.30 Two independent studies reported that exosomes derived from LPS-preconditioned dental follicle stem cells accelerated alveolar bone formation and periodontal tissue regeneration in experimental periodontitis.31,32 Having confirmed the uptake of exosomes by RAW264.7 cells, we continue to study the differential effects of NG-PDLSCs-Exo and HG-PDLSCs-Exo on osteoclast differentiation and periodontal tissue regeneration. Our results showed that NG-PDLSCs-Exo strongly inhibited osteoclast formation and differentiation in vitro and attenuated alveolar bone loss in the experimental periodontitis mice model compared to HG-PDLSCs-Exo.
We next examined the mechanisms underlying the various effects of the PDLSCs-Exo. Exosomes exert effects on repairing damaged tissues by transporting DNA, proteins, and miRNAs. Lai et al proposed that exosomes derived from bone marrow stem cells (BM-MSC-Exos) could deliver osteogenic miRNAs to target cells, thereby enhancing bone formation and ameliorating periodontitis-induced bone loss.33 Human periodontal ligament stem cells ameliorate periodontal inflammation by modulating the Th17/Treg balance through the release of miR-155-5p.34 Exosomes from human periodontal ligament stem cells reduce periodontal inflammation in rats by upregulating miR-205-5p.35 Our previous study revealed that PDLSCs (HG-PDLSCs) incubated in high glucose showed an impaired osteogenic differentiation ability compared to normal glucose-treated PDLSCs (NG-PDLSCs), possibly related to the upregulation of miR-31-5p expression in HG-PDLSCs.23 Recently, exosomal miR-31-5p was reported to inhibit osteoclastogenesis in the age-related bone marrow microenvironment.36 In combination with previous studies, we verified the high expression of miR-31-5p in HG-PDLSCs-Exo and proved that the effect of PDLSCs-Exo on osteoclast differentiation is related to miR-31-5p through RNA transfection.
MiRNAs are small non-coding RNAs involved in bone remodeling by regulating gene expression at the post-transcriptional level.37,38 It has been reported that miR-31-5p directly targets bone relevant markers such as SATB2, RUNX2 and OSX that regulate bone formation and regeneration.23,39,40 Deng et al found that RUNX2 binds directly to the miR-31-5p promoter and represses its transcription, while miR-31-5p negatively regulates SATB2 by targeting translational regulation in bone mesenchymal stem cells.39 Baglìo et al showed that miR-31-5p, one of the miRNAs differentially regulated during osteogenic differentiation of bone marrow-derived MSCs, controls the expression of Osterix through association with the 3’ untranslated region of these transcription facts.40 In this study, eNOS combined with bioinformatics analysis is a promising candidate for targeting the miR-31-5p gene. Furthermore, our experiments with luciferase reporter genes showed that miR-31-5p directly targets eNOS. The expression level of miR-31-5p negatively correlated with eNOS protein abundance. These results suggest that the high glucose microenvironment upregulates the expression of miR-31-5p in PDLSCs-Exo. High expression of exosomal miR-31-5p affects osteoclast differentiation, thus disrupting alveolar bone homeostasis and eventually leading to alveolar bone resorption.
Conclusion
Taken together, these results suggest that PDLSCs-derived exosomal miR-31-5p regulates alveolar bone regeneration by targeting eNOS. This study could provide new insights into the role of exosomal miRNAs in diabetic periodontitis. However, this is limited by the lack of a thorough study between miR-31-5p and osteoclast differentiation of macrophages in vitro. In future studies, we will further investigate the correlation between miR-31-5p and eNOS, and explore the involvement of miR-31-5p in alveolar bone homeostasis through in vivo experiments.
Abbreviations
Exo, exosomes; PDLSCs, periodontal ligament stem cells; PDLSCs-Exo, exosomes derived from periodontal ligament stem cells; NG-PDLSCs-Exo, exosomes derived from normal glucose-cultured periodontal ligament stem cells; HG-PDLSCs-Exo, exosomes derived from high-glucose-preconditioned periodontal ligament stem cells; TRAP, tartrate-resistant acid phosphatase; qRT-PCR, quantitative real time-polymerase chain reaction; eNOS, endothelial nitric oxide synthase; CP, chronic periodontitis; hPDLSCs-Exo, exosomes derived from human periodontal ligament stem cells; hASCs-Exo, exosomes derived from human adipose-derived stem cells; PGE2, prostaglandin E2; MSCs, mesenchymal stem cells; MSCs-Exo, exosomes derived from mesenchymal stem cells; DMEM, Dulbecco modified Eagle medium; DAPI, 4′,6-diamidino-2-phenylindole; TEM, transmission electron microscopy; α-MEM, alpha minimal essential medium; M-CSF, macrophage colony-stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SD, standard deviation; ANOVA, one-way analysis of variance; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; SHEDs, human exfoliated deciduous stem cells; BM-MSC-Exos, exosomes derived from bone marrow stem cells; H&E, hematoxylin and eosin; CEJ-ABC, cementoenamel junctions to the alveolar bone crest.
Acknowledgments
This project was partially funded by the National Natural Science Foundation of China (Grant Number: 81700981).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sanz M, Marco Del Castillo A, Jepsen S, et al. Periodontitis and cardiovascular diseases: consensus report. J Clin Periodontol. 2020;47(3):268–288. doi:10.1111/jcpe.13189
2. Loe H. Periodontal disease. The sixth complication of diabetes mellitus. Diabetes Care. 1993;16(1):329–334. doi:10.2337/diacare.16.1.329
3. Winning L, Patterson CC, Neville CE, et al. Periodontitis and incident type 2 diabetes: a prospective cohort study. J Clin Periodontol. 2017;44(3):266–274. doi:10.1111/jcpe.12691
4. Almeida Abdo J, Cirano FR, Casati MZ, et al. Influence of dyslipidemia and diabetes mellitus on chronic periodontal disease. J Periodontol. 2013;84(10):1401–1408. doi:10.1902/jop.2012.120366
5. Jimenez M, Hu FB, Marino M, et al. Type 2 diabetes mellitus and 20 year incidence of periodontitis and tooth loss. Diabetes Res Clin Pract. 2012;98(3):494–500. doi:10.1016/j.diabres.2012.09.039
6. Graves DT, Ding Z, Yang Y. The impact of diabetes on periodontal diseases. Periodontol. 2020;82(1):214–224. doi:10.1111/prd.12318
7. Zheng J, Chen S, Albiero ML, et al. Diabetes activates periodontal ligament fibroblasts via NF-κB in vivo. J DENT RES. 2018;97:580–588. doi:10.1177/0022034518755697
8. Tomokiyo A, Wada N, Maeda H. Periodontal ligament stem cells: regenerative potency in periodontium. Stem Cells Dev. 2019;28(15):974–985. doi:10.1089/scd.2019.0031
9. Zhao M, Dai W, Wang H, et al. Periodontal ligament fibroblasts regulate osteoblasts by exosome secretion induced by inflammatory stimuli. Arch Oral Biol. 2019;105:27–34. doi:10.1016/j.archoralbio.2019.06.002
10. Chen FM, Gao LN, Tian BM, et al. Treatment of periodontal intrabony defects using autologous periodontal ligament stem cells: a randomized clinical trial. Stem Cell Res Ther. 2016;7(1):33. doi:10.1186/s13287-016-0288-1
11. Ma Y, Ji Y, Zhong T, et al. Bioprinting-based PDLSC-ECM screening for in vivo repair of alveolar bone defect using cell-laden, injectable and photocrosslinkable hydrogels. ACS Biomater Sci Eng. 2017;3(12):3534–3545. doi:10.1021/acsbiomaterials.7b00601
12. Park JC, Kim JM, Jung IH, et al. Isolation and characterization of human periodontal ligament (PDL) stem cells (PDLSCs) from the inflamed PDL tissue: in vitro and in vivo evaluations. J Clin Periodontol. 2011;38(8):721–731. doi:10.1111/j.1600-051X.2011.01716.x
13. Wen S, Dooner M, Cheng Y, et al. Mesenchymal stromal cell-derived extracellular vesicles rescue radiation damage to murine marrow hematopoietic cells. Leukemia. 2016;30:2221–2231. doi:10.1038/leu.2016.107
14. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–289. doi:10.1146/annurev-cellbio-101512-122326
15. Hollý D, Klein M, Mazreku M, et al. Stem cells and their derivatives-implications for alveolar bone regeneration: a comprehensive review. Int J Mol Sci. 2021;22(21):11746. doi:10.3390/ijms222111746
16. Lei F, Li M, Lin T, et al. Treatment of inflammatory bone loss in periodontitis by stem cell-derived exosomes. Acta Biomater. 2022;141:333–343. doi:10.1016/j.actbio.2021.12.035
17. Zhao Y, Gong Y, Liu X, et al. The experimental study of periodontal ligament stem cells derived exosomes with hydrogel accelerating bone regeneration on alveolar bone defect. Pharmaceutics. 2022;14(10):2189. doi:10.3390/pharmaceutics14102189
18. Gao M, Gao W, Papadimitriou JM, et al. Exosomes-The enigmatic regulators of bone homeostasis. Bone Res. 2018;6:36. doi:10.1038/s41413-018-0039-2
19. Wu D, Chang X, Tian JJ, et al. Bone mesenchymal stem cells stimulation by magnetic nanoparticles and a static magnetic field: release of exosomal miR-1260a improves osteogenesis and angiogenesis. J Nanobiotechnology. 2021;19(1):209. doi:10.1186/s12951-021-00958-6
20. Chen S, Tang Y, Liu Y, et al. Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 2019;52(5):e12669. doi:10.1111/cpr.12669
21. Liu M, Chen R, Xu Y, et al. Exosomal miR-141-3p from PDLSCs alleviates high glucose-induced senescence of PDLSCs by activating the KEAP1-NRF2 signaling pathway. Stem Cells Int. 2023;2023:7136819. doi:10.1155/2023/7136819
22. Lin C, Yang Y, Wang Y, et al. Periodontal ligament fibroblasts-derived exosomes induced by PGE2 inhibit human periodontal ligament stem cells osteogenic differentiation via activating miR-34c-5p/SATB2/ERK. Exp Cell Res. 2022;419(2):113318. doi:10.1016/j.yexcr.2022.113318
23. Zhen L, Jiang X, Chen Y, et al. MiR-31 is involved in the high glucose-suppressed osteogenic differentiation of human periodontal ligament stem cells by targeting Satb2. Am J Transl Res. 2017;9(5):2384–2393.
24. Song C, Yang X, Lei Y, et al. Evaluation of efficacy on RANKL induced osteoclast from RAW264.7 cells. J Cell Physiol. 2019;234(7):11969–11975. doi:10.1002/jcp.27852
25. Andreev D, Liu M, Weidner D, et al. Osteocyte necrosis triggers osteoclast-mediated boneloss through macrophage-inducible C-type lectin. J Clin Invest. 2020;130(9):4811–4830. doi:10.1172/JCI134214
26. Shi N, Kong C, Yuan L, et al. The bidirectional relationship between periodontitis and diabetes: new prospects for stem cell-derived exosomes. Bio Pharmaco. 2023;165:115219. doi:10.1016/j.biopha.2023.115219
27. Wang M, Li J, Ye Y, et al. SHED-derived conditioned exosomes enhance the osteogenic differentiation of PDLSCs via Wnt and BMP signaling in vitro. Differentiation. 2020;111(111):1–11. doi:10.1016/j.diff.2019.10.003
28. Lai RC, Fu JH, Lim LP, et al. Mesenchymal stem cell exosomes enhance periodontal ligament cell functions and promote periodontal regeneration. Acta Biomat. 2019;89:252–264. doi:10.1016/j.actbio.2019.03.021
29. Lan Q, Xiao X, Bi X, et al. Effects of periodontal ligament stem cell-derived exosomes on osteoblastic proliferation, migration, differentiation, apoptosis, and signaling pathways. Oral Dis. 2022;10:1111.
30. Gao Y, Yuan Z, Yuan X, et al. Bioinspired porous microspheres for sustained hypoxic exosomes release and vascularized bone regeneration. Bioact Mater. 2022;14:377–388. doi:10.1016/j.bioactmat.2022.01.041
31. Shi W, Guo S, Liu L, et al. Small extracellular vesicles from lipopolysaccharide-preconditioned dental follicle cells promote periodontal regeneration in an inflammatory microenvironment. ACS Biomater Sci Eng. 2020;6(10):5797–5810. doi:10.1021/acsbiomaterials.0c00882
32. Huang Y, Liu Q, Liu L, et al. Lipopolysaccharide-preconditioned dental follicle stem cells derived small extracellular vesicles treating periodontitis via reactive oxygen species/mitogen-activated protein kinase signaling-mediated antioxidant effect. Int J Nanomedicine. 2022;17:799–819. doi:10.2147/IJN.S350869
33. Lai S, Deng L, Liu C, et al. Bone marrow mesenchymal stem cell-derived exosomes loaded with miR-26a through the novel immunomodulatory peptide DP7-C can promote osteogenesis. Biotechnol Lett. 2023;45(7):905–919. doi:10.1007/s10529-023-03376-w
34. Zheng Y, Dong C, Yang J, et al. Exosomal microRNA-155-5p from PDLSCs regulated Th17/ Treg balance by targeting sirtuin-1 in chronic periodontitis. J Cell Physiol. 2019;234(11):20662–20674. doi:10.1002/jcp.28671
35. Kang L, Miao Y, Jin Y, Shen S, Lin X. Exosomal miR-205-5p derived from periodontal ligament stem cells attenuates the inflammation of chronic periodontitis via targeting XBP1. Immun Inflamm Dis. 2023;11(1):e743. doi:10.1002/iid3.743
36. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi:10.1038/nrg2290
37. Gong K, Qu B, Liao D, et al. MiR-132 regulates osteogenic differentiation via downregulating Sirtuin1 in a peroxisome proliferator-activated receptor β/δ- dependent manner. Biochem Biophys Res Commun. 2016;478(1):260–267. doi:10.1016/j.bbrc.2016.07.057
38. Qadir AS, Um S, Lee H, et al. MiR-124 negatively regulates osteogenic differentiation and in vivo bone formation of mesenchymal stem cells. J Cell Biochem. 2015;116(5):730–742. doi:10.1002/jcb.25026
39. Deng Y, Zhou H, Zou D, et al. The role of miR31–modified adipose tissue–derived stem cells in repairing rat critical-sized calvarial defects. Biomaterials. 2013;34(28):6717–6728. doi:10.1016/j.biomaterials.2013.05.042
40. Baglìo SR, Devescovi V, Granchi D, et al. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013;527(1):321–331. doi:10.1016/j.gene.2013.06.021
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.