Back to Journals » OncoTargets and Therapy » Volume 16
Partial Response to Crizotinib in a Lung Adenocarcinoma Patient with a Novel FBXO11 (Intergenic)-ALK (Exon 20-29) Fusion
Authors He J, Yao Y, Quan F , Lu Z, Wang J , Gao W
Received 29 January 2023
Accepted for publication 13 June 2023
Published 7 July 2023 Volume 2023:16 Pages 535—540
DOI https://doi.org/10.2147/OTT.S406234
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Jing He,1,* Youyuan Yao,1,* Fei Quan,2 Zhongyu Lu,2 Jian Wang,1 Wen Gao1
1Medical Oncology Department, Jiangsu Province Hospital, Nanjing, Jiangsu, 210029, People’s Republic of China; 2The Medical Department, Jiangsu Simcere Diagnostics Co., Ltd, Nanjing, Jiangsu, 210021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wen Gao; Jian Wang, Medical Oncology Department, Jiangsu Province Hospital, 300 Guangzhou Road, Nanjing, Jiangsu, 210029, People’s Republic of China, Tel +86 025 68305755, Fax +86 025 68305758, Email [email protected]; [email protected]
Abstract: Intergenic-gene fusion detected by DNA-seq is particularly confusing for drug selection since the function of the intergenic region located upstream is unknown. We reported a case of a 49-year-old male with advanced lung adenocarcinoma, who was detected FBXO11 (intergenic)-ALK (exon 20-29) by DNA-seq, and FISH analysis revealed a positive result. The patient was treated with crizotinib and achieved a PR. The canonical EML4 (exon 1-13)-ALK (exon 20-29) fusion verified by RNA-seq suggested a complex EML4 (exon 1-13)-FBXO11 (intergenic)-ALK (exon 20-29) tripartite rearrangement at the DNA level. Our case emphasized the necessity of RNA-seq for verifying intergenic-gene fusion. Simultaneously, the pathogenic germline SLX4 variant and extensive CNVs of DNA segment were detected by DNA-seq deserves our attention.
Keywords: intergenic fusion, lung adenocarcinoma, ALK, SLX4, chromothripsis
Introduction
ALK rearrangement occurs in approximately 3~7% of non-small-cell lung cancer (NSCLC) patients.1 ALK fusion-positive patients acquired durable benefit from ALK tyrosine kinase inhibitors (TKIs),2 but relatively refractory to single-agent immune checkpoint inhibitors.3 Therefore, it is important to screen ALK fusion-positive tumors. However, traditional fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) cannot identify rare partners. Apart from EML4, an increasing number of gene fusion partners have been identified by DNA sequencing (DNA-seq). Moreover, multiple breakpoints have been reported, the introduction of RNA sequencing (RNA-seq) helps to distinguish whether a fusion at the DNA level forms a valid transcript. Besides the advantage in identifying complex fusions, the wider detection coverage of next-generation sequencing (NGS) allows it to provide additional information of concomitant mutations, which have been reported to impact prognostic of ALK-rearranged NSCLC.4 Above all, it is necessary to detect ALK rearrangement events accurately. Herein, we report a case of lung adenocarcinoma cancer who carried a rare intergenic-ALK fusion which confirmed to be a classical fusion by RNA-seq and FISH, the patient successfully benefited from crizotinib. Meanwhile, we posit that the molecular features and germline variant provided by DNA-seq will provide novel insight into complex fusions.
Case Presentation
A 49-year-old male was admitted for a dry cough that had become progressively worse for 2 months. On August 26, 2021, the enhanced computed tomography (CT) scan revealed a mass in the middle and lower lobe of the right lung with bilateral pulmonary multiple solid nodules and multiple enlarged lymph nodes in the mediastinum, bilateral hilum, left axilla, diaphragmatic angle and hepatogastric space. The right lung puncture biopsy immunohistochemistry showed CK7 (+), Napsin A (+), TTF-1 (+) and Ki67 (15%) (Figure 1A–D) revealed poorly differentiated lung adenocarcinoma. According to the TNM classification of the 8th edition of the Union of International Cancer Control (UICC), the patient was diagnosed with clinical stage IV (T4N3M1).
 |
Figure 1 Histological findings. (A) Hematoxylin and eosin (HE) staining. (B–D) IHC results of TTF-1, CK7 and Napsin A. |
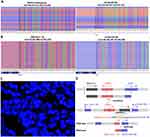
DNA-seq analysis of tumor tissue and plasma based on a 551-gene panel (SIMCEREDX Inc, Nanjing, China) revealed a novel FBXO11 (intergenic)-ALK (exon 20-29) fusion (Figure 2A), in addition, a SLX4 germline pathogenic variation and increased copy number of several genes were found. Considering that the function of the intergenic region is unknown, we used RNA-seq and FISH to verify and the corresponding results were EML4 (exon 1-13)-ALK (exon 20-29) fusion (Figure 2B) and ALK(+) (Figure 2C) respectively. Studies have shown that crizotinib significantly improved progression-free survival (PFS) and overall survival (OS) in ALK-positive patients compared with chemotherapy,5,6 which is recommended for first-line treatment in the NCCN guidelines. After full informed consent from the patient and his family, the patient was treated with crizotinib orally at a dose of 250mg daily on September 4, 2021, his cough was significantly relieved after a week and achieved continuous PR during the follow-up (Figure 3A–D).
 |
Figure 3 Representative clinical images before and after crizotinib treatment. (A–D) Comparison of primary lesions on baseline and every 8 weeks. |
Discussion
It is reported that approximately 25.5% of NSCLC patients identified as ALK, ROS1 or RET rearrangement-positive by DNA NGS were uncommon fusions, some of which neither a functional transcript nor a chimeric fusion protein was identified. Interestingly, among the patients verified positive by RNA NGS, genomic breakpoints and transcriptional-level breakpoints did not match in most cases.7,8 Intergenic region rearrangement, reciprocal rearrangement and noncanonical extron noncanonical are all likely to be identified as classical fusions at the transcript level.7,8 For our patient, it is difficult to determine whether it is an effective oncogenic fusion based on the DNA-seq results alone, because FBXO11 is a rare ALK fusion partner and the intergenic region is generally considered nonfunctional. Our case is an example for the potential significance of intergenic fusions and the importance of additional RNA-seq verification, which prevents ALK fusion-positive patients from missing out on targeted therapy and provides actual information about the fusion partner.
We are still puzzled by the inconsistency between DNA NGS and RNA NGS results. It is well known that intergenic region fusion upregulates downstream target genes mainly in two ways: reposition the upstream and downstream regulatory sequences or generate chimeric transcripts.9 RNA-seq showed that our patient matched the latter explanation. The types of alteration such as inversion, deletion, translocation and tandem duplication have been widely reported that can give rise to gene-intergenic fusions.9 Therefore, we make the following derivation according to the existing results. Firstly, considering ALK, EML4 and FBXO11 are all located on chromosome 2, but ALK and EML4 are transcribed in opposite directions, we speculated that inversion was necessary in the rearrangement process. Subsequently, the chromosome fragment between FBXO11 and EML4 (exon 1-13) was deleted, which produced FBXO11 (intergenic)-ALK (exon 20-29) fusion detected by DNA-seq. Finally, the intergenic region was cleaved during transcription to form EML4 (exon 1-13)-ALK (exon 20-29) fusion at the RNA level.10 It is worth noting that intergenic fusion alone was unlikely to be transcribed to EML4 (exon 1-13)-ALK (exon 20-29), there was more likely to be a complex tripartite rearrangement at the DNA level,8 constituting EML4 (exon 1-13), FBXO11 intergenic and ALK (exon 20-29) (Figure 2D). Unfortunately, the detection of long fusions containing intergenic regions is a difficult part of sequencing, the third long-read sequencing technology may solve ambiguous alignment of short read length,11 however, tissue samples taken at the patient’s initial visit have run out, retesting of resample will be considered after disease progressed. Our case suggests that the spread of third-generation sequencing technology could help reveal a more complete picture of genomic rearrangements, especially the complex rearrangements of long fragments.
What causes this complex fusion? It has been reported that chromothripsis and chromoplast are the main causes of complex genomic rearrangements,12 leading to intergenic region fusion detected by DNA-seq. Chromothripsis-like rearrangement may be the consequence of DNA repair disorders, resulting in chromosome instability.13 Genes involved in homologous recombination (HR) mediated DNA repair are worth more attention. Our patient carried a heterozygous pathogenic germline variant in SLX4 (c.2137C>T p.R713*). Liu et al reported somatic SLX4 mutations occurred in 9% of 44 ALK fusion positive Chinese NSCLC patients.14 SLX4 regulates three separate endonucleases and is involved in the repair of DNA crosslinks and the resolution of recombination intermediates,15 SLX4 is identified as an important regulator of DNA repair.16 Biallelic mutations in SLX4 are thought to be associated with Fanconi Anemia; nonetheless, heterozygous germline frame-shifting mutations in the SLX4 gene have been observed in individual patients, suggesting that it may cause partial protein truncation.17 Since SLX4 belongs to the HR pathway, we speculated that the impaired protein function of SLX4 might be related to chromothripsis. Moreover, the frequency of chromothripsis in lung adenocarcinomas was up to 40%, a hallmark of chromothripsis is loss of DNA fragments generated by the shattering of the DNA, resulting in copy number oscillations.18 The extensive copy number variations (CNVs) of DNA segment in the patient (Table 1) is concordant with that of the literature. The complex tripartite rearrangement may attribute to pathogenic germline variants in HR pathway genes, the CNVs of DNA fragments offer some support for this view; nevertheless, whole genome sequencing is needed for further verification. It is reported that about 5.91% of Chinese lung cancer patients could carry pathogenic/likely pathogenic germline mutations, which may promote tumorgenesis by inducing related mutation type.19 Our case shows that, compared with traditional FISH/IHC, NGS provides a better throughput and visualization, which contributes to a comprehensive understanding of the intergenic fusion tumors. The germline variant and extensive CNVs of DNA segment of the patient’s tumor distinguish it from the classic ALK fusion tumor. This leads us to speculate whether germline variants of HR pathway genes are more likely to induce complex fusions.
 |
Table 1 All Detected Variants in the Patient |
Does complex fusion affect drug efficacy compared to classical fusion? Cai et al revealed that there was no difference in PFS between patients with and without intergenic-ALK fusion in the patients who received first-line crizotinib.20 However, the conclusion was limited by the lack of RNA-seq validation. Moreover, 5’ partner may affect the biology of the fusion, and the distant promoter may also affect the concentration of transcript.21 As previously discussed, chromothripsis caused much of the complex rearrangement, which predicted a poor prognosis.18 In addition to oncogenic ALK fusion, the patient carried a pathogenic germline variant of HR pathway gene simultaneously. On the one hand, HR pathway gene mutations were associated with better immunotherapeutic effect, on the other hand, immune efficacy in ALK-fused patients was quite poor; therefore, whether HR pathways genes co-mutations could affect the efficacy of ALK-TKIs and immunotherapy remains to be explored.
Conclusions
We used NGS to identify a novel FBXO11 (intergenic)-ALK (exon 20-29) fusion that is actually a classical EML4-ALK fusion at the transcriptional level, which help a lung adenocarcinoma patient to benefit from crizotinib. Our case is an example for the potential significance of intergenic fusions and the importance of additional RNA-seq verification. In the meanwhile, DNA-seq revealed extensive CNVs of DNA segment and a pathogenic germline SLX4 variant, which provide new insights into intergenic fusions.
Abbreviations
NSCLC, non-small-cell lung cancer; TKIs, tyrosine kinase inhibitors; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; DNA-seq, DNA sequencing; RNA-seq, RNA sequencing; NGS, next-generation sequencing; CT, computed tomography; UICC, Union of International Cancer Control; PFS, progression-free survival; OS, overall survival; HR, homologous recombination; CNVs, copy number variations.
Data Sharing Statement
The original contributions presented in the study are included in the article, and further inquiries can be directed to the corresponding author.
Ethics Approval and Informed Consent
The authors declare that written informed consent was obtained from the patient’s and institutional approval for the publication of data and images. The research gene analysis was with ethics approval (HREC ID 4814).
Consent for Publication
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Ran Ding, Siqi Chen, Dr. Chuang Qi, Dr. Dongsheng Chen and Dr. Wanglong Deng from Jiangsu Simcere Diagnostics for their kind assistance. Jing He and Youyuan Yao are co-first authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Fei Quan and Zhongyu Lu were employed by Jiangsu Simcere Diagnostics Co., Ltd during the conduct of the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
2. Solomon BJ, Bauer TM, Mok TSK, et al. Efficacy and safety of first-line lorlatinib versus crizotinib in patients with advanced, ALK-positive non-small-cell lung cancer: updated analysis of data from the Phase 3, randomised, open-label CROWN study. Lancet Respir Med. 2023;11(4):354–366. doi:10.1016/S2213-2600(22)00437-4
3. Sankar K, Nagrath S, Ramnath N. Immunotherapy for ALK-rearranged non-small cell lung cancer: challenges inform promising approaches. Cancers. 2021;13(6):1476. doi:10.3390/cancers13061476
4. Li J, Zhang B, Zhang Y, et al. Concomitant mutation status of ALK-rearranged non-small cell lung cancers and its prognostic impact on patients treated with crizotinib. Transl Lung Cancer Res. 2021;10(3):1525–1535. doi:10.21037/tlcr-21-160
5. Solomon BJ, Kim D-W, Y-L W, et al. Final overall survival analysis from a study comparing first-line crizotinib versus chemotherapy in ALK-mutation-positive non–small-cell lung cancer. J Clin Oncol. 2018;36(22):2251–2258. doi:10.1200/JCO.2017.77.4794
6. Solomon BJ, Mok T, Kim D-W, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi:10.1056/NEJMoa1408440
7. Li W, Guo L, Liu Y, et al. Potential unreliability of uncommon ALK, ROS1, and RET genomic breakpoints in predicting the efficacy of targeted therapy in NSCLC. J Thorac Oncol. 2021;16(3):404–418. doi:10.1016/j.jtho.2020.10.156
8. Li W, Liu Y, Li W, Chen L, Ying J. Intergenic breakpoints Identified by DNA sequencing confound targetable kinase fusion detection in NSCLC. J Thorac Oncol. 2020;15(7):1223–1231. doi:10.1016/j.jtho.2020.02.023
9. Yun JW, Yang L, Park HY, et al. Dysregulation of cancer genes by recurrent intergenic fusions. Genome Biol. 2020;21(1):166. doi:10.1186/s13059-020-02076-2
10. Xiang C, Guo L, Zhao R, et al. Identification and validation of noncanonical RET fusions in non–small-cell lung cancer through DNA and RNA sequencing. J Mol Diagn. 2022;24(4):374–385. doi:10.1016/j.jmoldx.2021.12.004
11. Xu L, Wang X, Lu X, et al. Long-read sequencing identifies novel structural variations in colorectal cancer. PLoS Genet. 2023;19(2):e1010514. doi:10.1371/journal.pgen.1010514
12. Lee JK, Park S, Park H, Kim S, Kim YT. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell. 2019;177(7):1842–1857.e21. doi:10.1016/j.cell.2019.05.013
13. Nazaryan-Petersen L, Bjerregaard VA, Nielsen FC, Tommerup N, Tümer Z. Chromothripsis and DNA repair disorders. J Clin Med. 2020;9(3):613. doi:10.3390/jcm9030613
14. Liu S, Huang T, Liu M, et al. The genomic characteristics of ALK fusion positive tumors in Chinese NSCLC patients. Front Oncol. 2020;10:726. doi:10.3389/fonc.2020.00726
15. Young SJ, West SC. Coordinated roles of SLX4 and MutSβ in DNA repair and the maintenance of genome stability. Crit Rev Biochem. 2021;56(2):157–177. doi:10.1080/10409238.2021.1881433
16. Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43(2):138–141. doi:10.1038/ng.751
17. Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43(2):142–146. doi:10.1038/ng.750
18. Cortés-Ciriano I, Lee JJ, Xi R, et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat Genet. 2020;52(3):331–341. doi:10.1038/s41588-019-0576-7
19. Peng W, Li B, Li J, et al. Clinical and genomic features of Chinese lung cancer patients with germline mutations. Nat Commun. 2022;13(1):1268. doi:10.1038/s41467-022-28840-5
20. Cai C, Tang Y, Li Y, et al. Distribution and therapeutic outcomes of intergenic sequence-ALK fusion and coexisting ALK fusions in lung adenocarcinoma patients. Lung Cancer. 2021;152:104–108. doi:10.1016/j.lungcan.2020.12.018
21. Li Y, Duan P, Guan Y, et al. High efficacy of alectinib in a patient with advanced lung adenocarcinoma with 2 rare ALK fusion sites: a case report. Transl Lung Cancer Res. 2022;11(1):100–110. doi:10.21037/tlcr-21-1039
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.