Back to Journals » Drug Design, Development and Therapy » Volume 11
Paeoniflorin suppresses pancreatic cancer cell growth by upregulating HTRA3 expression
Authors Li YJ, Gong LL, Qi RL, Sun Q, Xia XX, He HH, Ren JS, Zhu ON, Zhuo DB
Received 19 April 2017
Accepted for publication 29 June 2017
Published 23 August 2017 Volume 2017:11 Pages 2481—2491
DOI https://doi.org/10.2147/DDDT.S134518
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Yuejun Li,1,* Lili Gong,2,* Ruili Qi,2 Qian Sun,2 Xinxin Xia,3 Haihui He,1 Jianshu Ren,1 Ouning Zhu,1 Debin Zhuo1
1The Third Department of Oncology, The First Affiliated Hospital of Hunan College of Traditional Chinese Medicine, Zhuzhou, Hunan, 2State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-Sen University, Guangzhou, Guangdong, 3Department of Traditional Chinese Medicine, The First Affiliated Hospital of Xian Jiaotong University, Xian, Shanxi, People’s Republic of China
*These authors contributed equally to this work
Abstract: Paeoniflorin (PF) is an active monoterpene glycoside extracted from Paeonia lactiflora Pall. PF has exhibited antitumor effects in various cancer types. However, the effects of PF in pancreatic cancer are largely unexplored. Here, we showed that PF suppressed growth of pancreatic cancer cell lines Capan-1 and MIAPaCa-2 and profoundly sensitized these cells to X-ray irradiation. Through microarray analysis, we identified HTRA3, a tumor-suppressor candidate gene, as the most increased gene upon PF treatment in Capan-1 cells. Ectopic expression of HTRA3 led to reduced cell proliferation and increased expression of apoptotic protein Bax, suggesting a tumor suppressive role of HTRA3 in pancreatic cancer cells. Together, our results provide a set group of genetic proofs and biological proofs that PF inhibited pancreatic cancer growth by upregulating HTRA3.
Keywords: traditional Chinese medicine, X-ray irradiation, colony formation assay, microarray analysis, cancer inhibition
Introduction
Pancreatic cancer is currently the fourth most common cause of cancer death, and it is expected to rise to the second most common cause of death by 2030.1,2 The overall median survival of pancreatic cancer is <1 year from diagnosis, highlighting the need for the development of new therapeutic strategies.
Paeoniflorin (PF) is an active component of Paeonia lactiflora Pall (Ranunculaceae family). It has been recently demonstrated that this component has antitumor effects in multiple cancer types, such as glioma, breast, gastric, colorectal, cervical and lung cancers.3–8 PF also prevents hypoxia-induced epithelial–mesenchymal transition (EMT) in breast cancer cells and macrophage-mediated lung cancer metastasis.9,10 However, whether PF has any effect on pancreatic cancer is largely unknown.
HTRA3 belongs to the highly conserved HtrA family of stress-related serine proteases. It was initially identified as a serine protease in the developing placenta.11,12 Dysregulation of HTRA3 has been reported in different types of cancer.13–17 Particularly, downregulation of HTRA3 was found to be associated with the progression of endometrial and ovarian cancers.14,15 In lung cancer cells, HTRA3 was released from mitochondrial to cytosol upon etoposide or cisplatin treatments, promoting cytotoxicity induced by those agents.18 Therefore, HTRA3 is proposed as a tumor suppressor and a potential therapeutic target in cancer.19,20
In the present study, we showed that PF treatment led to increased cell apoptosis and decreased colony formation in pancreatic cancer cell lines Capan-1 and MIAPaCa-2. We conducted microarray analysis and identified HTRA3 as the most unregulated gene with PF treatment in Capan-1 cells. We further confirmed elevated HTRA3 mRNA and protein expression upon PF treatment. Ectopic expression of HTRA3 inhibited proliferation and induced Bax expression in Capan-1 cells. Together, these results demonstrate that PF inhibited pancreatic cancer cell growth by upregulating HTRA3 expression and promoting HTRA3-mediated apoptosis.
Materials and methods
Cell culture and chemicals
Capan-1 and MIAPaCa-2 cell lines were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). Both cell lines were maintained in Dulbecco’s modification of Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. PF was purchased from Selleckchem (Houston, TX, USA). FLAG-HTRA3 plasmid was purchased from YouBio (Changsha, People’s Republic of China) and verified by DNA sequencing. Antibodies against HTRA3, Bax, GAPDH, and β-actin were purchased from Sangon Biotech (Shanghai, People’s Republic of China). A PE Annexin V apoptosis detection kit was purchased from BD Biosciences (San Jose, CA, USA; BD Pharmingen™, # 559763), and apoptosis assay was performed following the manufacturer’s procedure. The transfection reagent Lipofectamine 3000 was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Clonogenic cell survival assay
Capan-1 and MIAPaCa-2 cells were seeded in six-well plates at 200 cells/well. The indicated concentrations of PF were added the next day, and cells were left for 7 days to form colonies. Colonies were stained with 0.25% crystal violet and 25% methanol in phosphate buffered saline solution for visualization. For Capan-1 cells, which generally form tight and distinct colonies, colonies with ≥50 cells were counted manually and digitally using the ImageJ software (National Institute of Mental Health, Betheseda, MD, USA) with customized parameters that were optimized on the basis of three preliminary manual counts. However, MIAPaCa-2 cells exhibited a more scattered pattern, which made it hard to determine the colony number. Therefore, their colony area was measured instead using the “colony area” plugin of ImageJ software.21 The relative colony area was calculated by multiplying the colony area to the colony intensity.
X-ray irradiation
Cells plated on six-well plate were pretreated with PF for 24 h before irradiation. The indicated doses of X-ray radiation were generated by RS2000 irradiator (Rad Source Technologies, Suwanee, GA, USA). After irradiation, cells were recultured for 7 days before clonogenic assay.
Cell proliferation assay
Cell proliferation was measured using the PrestoBlue® Cell Viability Reagent (# A13261; Thermo Fisher Scientific). Cells for each experimental condition were seeded in triplicate in six-well plates at 200 cells/well. The next day, cells were treated with dimethyl sulfoxide (DMSO) or the indicated concentrations of PF. After 7 days, PrestoBlue Cell Viability Reagent was added to each well and incubated for 1 h at 37°C. Then, the fluorescence was measured by a Synergy™ H1 Multi-Mode Reader (BioTek, Winooski, VT, USA). After background subtraction, the cell viability was calculated as the percentage change relative to the control cells.
Microarray analysis
Capan-1 cells were treated with PF or DMSO for 48 h and then lysed/stored in TRIzol™ reagent (# 15596018; Thermo Fisher Scientific) before sending to CapitalBio Technology Company (Beijing, People’s Republic of China) for microarray analysis. Briefly, cRNA was prepared by MessageAmp™ Premier RNA Amplification Kit (AM1792) and hybridized to GeneChip® PrimeView™ Human Gene Expression Array. Data were normalized using Robust Multichip Average (RMA) and log-2 transformed. Gene expression differences were considered significant if P<0.05. Heatmap was generated from normalized microarray data using Cluster 3.0. Functional enrichment analysis was performed using FunRich software (http://www.funrich.org)22 and DAVID (Bioinformatics Resources v6.7). Raw data have been submitted to the GEO database under accession number GSE97124.
Quantitative reverse-transcription (qRT) polymerase chain reaction (PCR) analysis
Selected microarray results were validated by qRT-PCR analysis. Briefly, reverse transcription was conducted using an RevertAid First Strand cDNA Synthesis Kit (# K1622; Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time PCR was conducted on the LightCycler 480 II using SYBR® Green Real-Time PCR Master Mix (TIANGEN Biotech, Beijing, People’s Republic of China). Primers used for qRT-PCR are as follows:
HTRA3: 5′-TGGCTTCATCATGTCAGAGG-3′, 5′-GGCAATGTCCGACTTCTTGT-3′ |
HMOX1: 5′-CTTCTTCACCTTCCCCAACA-3′, 5′-GCTCTGGTCCTTGGTGTCAT-3′ |
NRCAM: 5′-CCCAATTGGATTACCACCAC-3′, 5′-CTCTGGGAGGACATTGGAAA-3′ |
KATNBL1: 5′-CTCCAAAACAGTTGGCTGCT-3′, 5′-CAAGGATTTGGAAAGGGATG-3′ |
SERPINB3: 5′-GAAGATCGCCAACAAGCTCT-3′, 5′-GTTTGACTTTCCACCCAGGA-3′ |
β-Actin: 5′-TCACCAACTGGGACGACAT-3′, 5′-ATCTGGGTCATCTTCTCGC-3′ |
Transfection and Western blot analysis
Capan-1 cells seeded in six-well plate were transfected with indicated concentrations of FLAG-HTRA3 plasmid using Lipofectamine 3000 (# L3000008). After 48 h, total proteins were extracted using radio immuno precipitation assay lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 2 mM ethylene diamine tetraacetic acid, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate) in the presence of proteinase inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA). Equal amounts of proteins (30 μg) were separated by 10% dodecyl sulfate, sodium salt -polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The chemiluminescent signals were detected and captured by Tanon 5200.
Immunohistochemistry
Tissue arrays were obtained from US Biomax (# PA483C; Rockville, MD, USA). The deparaffinization and antigen retrieval were performed according to the manufacturer’s instruction. The slides were blocked in 5% goat serum for 20 min before incubating with anti-HTRA3 (# D161337, 1:100 dilution) for 2 h at room temperature. The immunohistochemistry staining procedure was carried out using the GTVision™ III System according to the manufacturer’s protocol (# GK500705; GeneTech, Shanghai, People’s Republic of China). The slides were counterstained with hematoxylin before observation by light microscopy with LEICA DM4000.
Statistical analysis
The statistical significance of differences between groups was determined by the two-tailed t-test. P<0.05 was considered as statistically significant.
Results
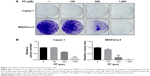
PF inhibited colony formation of pancreatic cancer cells
We first determined the effects of PF on pancreatic cancer cell growth by clonogenic cell survival assay. As shown in Figure 1, PF significantly reduced colony formation in both Capan-1 (Figure 1A, upper panel) and MIAPaCa-2 cells (Figure 1A, lower panel) in a dose-dependent manner. Treatment of 500 μM PF alone for 7 days led to ~40 and 80% inhibition in the growth of Capan-1 and MIAPaCa-2 cells, respectively (Figure 1B).
In clinical practice of pancreatic cancers, radiation therapy is usually used after surgery as adjuvant treatment, or in conjunction with chemotherapy for unresectable pancreatic cancers.
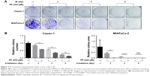
Therefore, we sought to determine whether PF could radio-sensitize pancreatic cancer cells.
Capan-1 and MIAPaCa-2 cells were subjected to 2–8 Gy X-ray irradiation (IR) before treatment of 500 μM PF or DMSO (vehicle control) (Figure 2A). We found that PF significantly enhanced the sensitivity of Capan-1 cells to IR, resulting in 60%, 51% and 12% more inhibition upon 2, 4, and 8 Gy IR exposure, respectively, as compared with IR alone (Figure 2B, left panel). Consistently, PF dramatically sensitized MIAPaCa-2 cells to IR, leading to a >60-fold decrease in colony formation as compared with IR alone (Figure 2B, right panel). Together, we showed that PF inhibited the proliferation of pancreatic cancer cells. Furthermore, addition of PF significantly enhanced the cytotoxic effects of IR, suggesting a potential role of PF as a radio-sensitizer in pancreatic cancer.
PF increased cell apoptosis in pancreatic cancer cells
Next, cell proliferation assay was performed to further determine the effects of PF on pancreatic cancer cell growth. PF inhibited cell growth in both pancreatic cancer cell lines. The half maximal (50%) inhibitory concentration (IC50) values for PF in Capan-1 and MIAPaCa-2 were 862.7 and 489.5 μM, respectively (Figure 3). In addition, PF treatment increased the percentage of apoptotic cells in both Capan-1 and MIAPaCa-2 cells, as determined by the 7-AAD/PE Annexin V apoptotic analysis (Figure 4A). Additionally, we found increased proapoptotic Bax protein levels when Capan-1 (Figure 4B) and MIAPaCa-2 (Figure 4C) cells were exposed to PF treatment.
PF treatment induced HTRA3 expression in Capan-1 cells
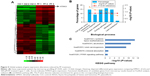
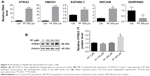
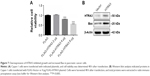
Next, we searched for the underlying mechanisms that are responsible for PF-induced suppression of pancreatic cancer cell growth. To obtain a broad molecular understanding of this process, we applied a genome-wide expression profiling approach to systematically analyze cellular transcriptome alterations in Capan-1 cells. Upon 500 μM PF treatment for 48 h, we identified a total of 77 reference genes that showed >1.5-fold change relative to the control group (34 upregulated genes and 43 downregulated genes) (P<0.05; Figure 5A). Functional enrichment analysis indicated that these altered genes were enriched for cell growth/maintenance, immune response, protein metabolism and so on (Figure 5B). Analysis of the altered genes by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database revealed involvement in systemic lupus erythematosus, alcoholism, and viral carcinogenesis signaling pathway (Figure 5C). Among the altered genes in response to PF treatment, we found that HTRA3 was the most significantly increased gene (Figure 5A). HTRA3 was initially discovered as a pregnancy-related serine protease to be upregulated in mouse uterus coinciding with placentation.11 Recently, HTRA3 was found to be implicated as a tumor suppressor by inhibiting cell invasion.20,23 In lung cancer cell lines, HTRA3 promoted etoposide- and cisplatin-induced cytotoxicities in lung cancer cell lines.18 Hence, HTRA3 might be a key molecule mediating PF-induced growth inhibition and apoptosis in pancreatic cancer cells. Consistent with our microarray data, we confirmed with qRT-PCR that Capan-1 cells exposed to PF had significantly increased HTRA3 mRNA than control cells (P<0.0001; Figure 6A). In addition, significantly increased HTRA3 protein levels were observed in response to 500 μM PF treatment as compared with DMSO treatment in Capan-1 cells (P<0.05; Figure 6B and C). However, we did not observe significant HTRA3 RNA or protein change in MIA PaCa-2 cells (Figure S1), suggesting that PF inhibits Capan-1 and MIAPaCa-2 through different mechanisms.
HTRA3 inhibited Capan-1 cell growth
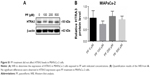
To the best of our knowledge, the effect of HTRA3 in pancreatic cancer has not been explored before. Here, we overexpressed HTRA3 in Capan-1 cells and showed that HTRA3 dose dependently led to significantly reduced cell viability (Figure 7A). Consistent with the inhibition of cell growth, increased protein level of Bax was detected upon HTRA3 overexpression (Figure 7B). Therefore, HTRA3 could possibly suppress the proliferation of pancreatic cancer cells via inducing the expression of proapoptotic Bax protein.
HTRA3 expression decreased in pancreatic cancer samples
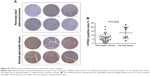
We studied the clinical relevance of HTRA3 in human cancers. Immunohistochemistry staining of HTRA3 was conducted on pancreatic cancer tissue microarrays (Figure 8). Expression of HTRA3 was compared in 31 individual pancreas duct adenocarcinomas and eight normal pancreatic tissues (Table S1). In normal pancreas tissue, HTRA3 generally displayed strong positive staining in the cytoplasm, whereas in cancer samples, HTRA3 is decreased (Figure 8A). Further quantification of the HTRA3-positive staining area revealed about twofold decreased HTRA3 signal in cancer samples as compared with normal tissue (Figure 8B). Together, these data suggest that HTRA3 expression is clinically significant in pancreatic cancer.
Discussion
In this study, we showed that PF significantly inhibited cell growth and sensitized cells to irradiation in pancreatic cancer. PF is the principal compound of “Radix Paeoniae Alba” that is commonly included in many traditional prescriptions to treat digestive system diseases for its anti-inflammatory and antipathogenic microorganism effects.24–26 PF has been recently shown to reduce cell viability in gastric cancer cells,5 glioma cells,3 ovarian cancer cells, and breast cancer cells.4 During the preparation of this manuscript, Hao et al27 reported that PF sensitized erlotinib-induced inhibition of cell viability in ErbB3-expressing pancreatic cancer cell lines, which relied on the expression of ErbB3. In their studies, up to 50 μM PF showed no effects on growth, colony formation, or apoptosis in ErbB3-deficient MIAPaCa-2.27 Here, we also found that PF <50 μM did not affect the growth of either MIAPaCa-2 or Capan-1 cells. The IC50 value of PF was 489.5 and 862.7 μM in MIAPaCa-2 and Capan-1 cells, respectively (Figure 3). Such differences may have resulted from the markedly differential proliferation rate of the two cell lines, with a doubling time of 40 h in MIAPaCa-2 and 96 h in Capan-1 cells.28,29
For the first time, we have systematically explored the molecular mechanism underlying PF-inhibited pancreatic cancer proliferation through transcriptome analysis upon PF treatment in Capan-1 cells. Among all the altered genes, HTRA3 was most significantly increased (Figure 5A). HTRA3 belongs to the HtrA family of stress-related serine proteases. HTRA3 sensitizes lung cancer cells to etoposide and cisplatin, where it may act as a mitochondrial cell death effector, suggesting its role as a tumor suppressor.18 Our data showed that overexpression of HTRA3 significantly upregulated Bax and suppressed cell viability in Capan-1 (Figure 7) and that decreased HTRA3 was associated with human pancreas carcinogenesis (Figure 8), clearly proving that HTRA3 is proapoptotic and tumor suppressing in pancreatic cancer cells.
PF has been recently shown to modulate multiple signaling pathways in different disease models. Gene ontology analysis of our microarray data revealed that genes significantly altered by PF treatment were involved in inflammatory response, cell proliferation, and response to hypoxia. The effects of PF on these biological processes have been well documented elsewhere. For example, the anti-inflammatory functions of PF were reported in studies on brain,25 spleen,30 and kidney.24 In breast cancer cells, PF was found to prevent hypoxia-induced EMT through suppressing HIFα expression.10 It is intriguing to explore whether such events may also contribute to PF-inhibited pancreatic cancer cell viability.
Conclusion
Together, we have shown the anticancer effects of PF in pancreatic cancer cell lines. Through a genome-wide transcriptome analysis, we found that PF increased HTRA3 expression, a previously uncharacterized mechanism by which PF suppressed proliferation and enhanced apoptosis in pancreatic cancer cell. Our findings not only revealed the therapeutic potential of PF in pancreatic cancer treatment but also demonstrated a novel proapoptotic role of HTRA3 in pancreatic cancer cells via inducing Bax.
Acknowledgment
This work was supported by the Hunan Administration Traditional Chinese Medicine Foundation (201737) and the National Science Foundation of China (81500738).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. | ||
Nie XH, Ou-yang J, Xing Y, et al. Paeoniflorin inhibits human glioma cells via STAT3 degradation by the ubiquitin-proteasome pathway. Drug Des Devel Ther. 2015;9:5611–5622. | ||
Zhang Q, Yuan Y, Cui J, Xiao T, Jiang D. Paeoniflorin inhibits proliferation and invasion of breast cancer cells through suppressing Notch-1 signaling pathway. Biomed Pharmacother. 2016;78:197–203. | ||
Zheng YB, Xiao GC, Tong SL, et al. Paeoniflorin inhibits human gastric carcinoma cell proliferation through up-regulation of microRNA-124 and suppression of PI3K/Akt and STAT3 signaling. World J Gastroenterol. 2015;21(23):7197–7207. | ||
Wang H, Zhou H, Wang CX, et al. Paeoniflorin inhibits growth of human colorectal carcinoma HT 29 cells in vitro and in vivo. Food Chem Toxicol. 2012;50(5):1560–1567. | ||
Zhang L, Zhang S. Modulating Bcl-2 family proteins and caspase-3 in induction of apoptosis by paeoniflorin in human cervical cancer cells. Phytother Res. 2011;25(10):1551–1557. | ||
Hung JY, Yang CJ, Tsai YM, Huang HW, Huang MS. Antiproliferative activity of paeoniflorin is through cell cycle arrest and the Fas/Fas ligand-mediated apoptotic pathway in human non-small cell lung cancer A549 cells. Clin Exp Pharmacol Physiol. 2008;35(2):141–147. | ||
Wu Q, Chen GL, Li YJ, Chen Y, Lin FZ. Paeoniflorin inhibits macrophage-mediated lung cancer metastasis. Chin J Nat Med. 2015;13(12):925–932. | ||
Zhou Z, Wang S, Song C, Hu Z. Paeoniflorin prevents hypoxia-induced epithelial–mesenchymal transition in human breast cancer cells. Onco Targets Ther. 2016;9:2511–2518. | ||
Nie GY, Li Y, Minoura H, et al. A novel serine protease of the mammalian HtrA family is up-regulated in mouse uterus coinciding with placentation. Mol Hum Reprod. 2003;9(5):279–290. | ||
Nie GY, Hampton A, Li Y, Findlay JK, Salamonsen LA. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem J. 2003;371(pt 1):39–48. | ||
Beleford D, Liu Z, Rattan R, et al. Methylation induced gene silencing of HtrA3 in smoking-related lung cancer. Clin Cancer Res. 2010;16(2):398–409. | ||
Bowden MA, Di Nezza-Cossens LA, Jobling T, Salamonsen LA, Nie G. Serine proteases HTRA1 and HTRA3 are down-regulated with increasing grades of human endometrial cancer. Gynecol Oncol. 2006;103(1):253–260. | ||
Narkiewicz J, Klasa-Mazurkiewicz D, Zurawa-Janicka D, Skorko-Glonek J, Emerich J, Lipinska B. Changes in mRNA and protein levels of human HtrA1, HtrA2 and HtrA3 in ovarian cancer. Clin Biochem. 2008;41(7–8):561–569. | ||
Zhao M, Ding JX, Nie GY, et al. HTRA3 is reduced in ovarian cancers regardless of stage. Cancer Invest. 2014;32(9):464–469. | ||
Bowden MA, Drummond AE, Fuller PJ, Salamonsen LA, Findlay JK, Nie G. High-temperature requirement factor A3 (Htra3): a novel serine protease and its potential role in ovarian function and ovarian cancers. Mol Cell Endocrinol. 2010;327(1–2):13–18. | ||
Beleford D, Rattan R, Chien J, Shridhar V. High temperature requirement A3 (HtrA3) promotes etoposide- and cisplatin-induced cytotoxicity in lung cancer cell lines. J Biol Chem. 2010;285(16):12011–12027. | ||
Zurawa-Janicka D, Skorko-Glonek J, Lipinska B. HtrA proteins as targets in therapy of cancer and other diseases. Expert Opin Ther Targets. 2010;14(7):665–679. | ||
Chien J, Campioni M, Shridhar V, Baldi A. HtrA serine proteases as potential therapeutic targets in cancer. Curr Cancer Drug Targets. 2009;9(4):451–468. | ||
Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One. 2014;9(3):e92444. | ||
Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. | ||
Singh H, Makino SI, Endo Y, Nie G. Inhibition of HTRA3 stimulates trophoblast invasion during human placental development. Placenta. 2010;31(12):1085–1092. | ||
Liu Q, Lin X, Li H, et al. Paeoniflorin ameliorates renal function in cyclophosphamide-induced mice via AMPK suppressed inflammation and apoptosis. Biomed Pharmacother. 2016;84:1899–1905. | ||
Gu X, Cai Z, Cai M, et al. Protective effect of paeoniflorin on inflammation and apoptosis in the cerebral cortex of a transgenic mouse model of Alzheimer’s disease. Mol Med Rep. 2016;13(3):2247–2252. | ||
Zhang HR, Peng JH, Cheng XB, Shi BZ, Zhang MY, Xu RX. Paeoniflorin atttenuates amyloidogenesis and the inflammatory responses in a transgenic mouse model of Alzheimer’s disease. Neurochem Res. 2015;40(8):1583–1592. | ||
Hao J, Yang X, Ding XL, et al. Paeoniflorin potentiates the inhibitory effects of erlotinib in pancreatic cancer cell lines by reducing ErbB3 phosphorylation. Sci Rep. 2016;6:32809. | ||
Kyriazis AP, Kyriazis AA, Scarpelli DG, Fogh J, Rao MS, Lepera R. Human pancreatic adenocarcinoma line Capan-1 in tissue culture and the nude mouse: morphologic, biologic, and biochemical characteristics. Am J Pathol. 1982;106(2):250–260. | ||
Yunis AA, Arimura GK, Russin DJ. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. Int J Cancer. 1977;19(1):128–135. | ||
Zhang H, Qi Y, Yuan Y, et al. Paeoniflorin ameliorates experimental autoimmune encephalomyelitis via inhibition of dendritic cell function and Th17 cell differentiation. Sci Rep. 2017;7:41887. |
Supplementary materials
  | Table S1 Patient and normal tissue information and the percentage of HTRA3-positive staining area |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.