Back to Journals » Journal of Inflammation Research » Volume 15
P2X7 Receptor and Heart Function in a Mouse Model of Systemic Inflammation Due to High Fat Diet
Authors Raggi F, Rossi C , Faita F , Distaso M , Kusmic C, Solini A
Received 7 January 2022
Accepted for publication 9 March 2022
Published 14 April 2022 Volume 2022:15 Pages 2425—2439
DOI https://doi.org/10.2147/JIR.S356038
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Francesco Raggi,1 Chiara Rossi,1 Francesco Faita,2 Mariarosaria Distaso,1 Claudia Kusmic,2 Anna Solini1
1Department of Surgical, Medical, Molecular and Critical Area Pathology, University of Pisa, Pisa, Italy; 2Institute of Clinical Physiology, Italian National Research Council, Pisa, Italy
Correspondence: Anna Solini; Francesco Raggi, Department of Surgical Medical Molecular and Critical Area Pathology, University of Pisa, Via Roma 67, Pisa, I-56126, Italy, Tel +39-50-993482 ; +39-50-992861, Fax +39-50-553235, Email [email protected]; [email protected]
Purpose: Low-grade inflammation contributes to heart failure in obesity or type 2 diabetes mellitus. The P2X7 receptor (P2X7R) is a key regulator of several pro-inflammatory responses in multiple tissues and organs; however, its involvement in the onset of heart dysfunction remains unclear. The study evaluated the role of P2X7R as a cardiac function regulator in C57BL/6J wild-type (WT) and P2X7R knockout (KO) mice by inducing systemic inflammation with high fat diet (HFD).
Methods: Specific parameters of systolic and diastolic function and heart morphology were measured in vivo before animal sacrifice by high-frequency ultrasonographic analysis. Gene and protein expression of cardiac biomarkers associated with inflammatory-oxidative pathways were evaluated by real-time PCR and Western Blotting.
Results: P2X7R-mediated up-regulation of the NLRP3-caspase-1 complex, increased expression of key oxidative stress (NOS-2, TNFα), and chemotactic (MCP-1) mediators were revealed in WT-HFD animals. In KO-HFD mice, such inflammatory-oxidative pathway was silent. Nevertheless, HFD induced in vivo a clear alteration of diastolic pattern (E/A: p < 0.03 vs WT-HFD) and a cardiac morphologic remodelling (left ventricular mass: p < 0.05 vs WT-HFD) only in P2X7R KO animals. Surprisingly, the transcriptional and protein expression of IL-1β and IL-6, usually regulated through P2X7R activation, were significantly higher in KO-HFD than in WT-HFD mice (both p < 0.05). Furthermore, an up-regulation of miR-214 and a down-regulation of miR-126 in heart of HFD-KO mice were observed, suggesting a link between such epigenetic dysregulation and cytokine overexpression as a potential pathophysiologic mechanism concurring to the progressive cardiac dysfunction.
Conclusion: These findings seem to suggest a cardioprotective role of P2X7R toward this tissue-specific inflammatory damage, likely through tissue homeostasis and organ functionality preservation.
Keywords: low-grade inflammation, high fat diet, diastolic dysfunction, P2X7 receptor
Introduction
Low-grade chronic inflammation plays a crucial role in the pathogenesis of atherosclerosis and insulin resistance, leading mechanisms in the development of cardiovascular disease (CVD) in individuals with obesity of type 2 diabetes.1,2 Subclinical inflammation is linked to an increased expression of genes related to oxidative stress, cytokine production and angiogenesis in visceral adipose tissue of these patients,3 impairing arteriolar function and endothelial vasodilatation with, eventually, endothelial dysfunction.4 Among several other factors, dietary habits might influence such risk: a high content of saturated fats is a major determinant of increased LDL-cholesterol levels, accelerated atherosclerosis and enhanced CVD incidence,5 while increasing evidence suggest as low-fat diets or high intake of polyphenol-rich food are related to a reduction of chronic degenerative diseases and mortality risk.6 However, diet affects not only the vasculature, but also the myocardial function.7,8 This epidemiologic evidence in humans is well mirrored by several experimental models: for example, a clear increase in the level of circulating inflammatory factors has been detected in both rat and mouse models of high-fat diet (HFD)-induced obesity,9,10 associated with evident structural and functional alterations of the heart.
The P2X7 purinergic receptor (P2X7R) modulates the assembly of the NLR Family Pyrin Domain Containing 3 (NLRP3) inflammasome,11 leading to the secretion of proinflammatory cytokines like interleukin (IL)-1β, IL-18 and IL-6 in several cellular and tissue structures.12,13 In the heart, such pathway is critical for high glucose-induced collagen synthesis and cardiac fibrosis, and its induction in an animal model of acute myocardial infarction leads to a significant increase in caspase-1 activity and enhanced cell death.14,15 Growing evidence show that microRNAs (miRNAs) are directly involved in the progression of several cardiovascular disorders, as they regulate the expression of genes implicated in the cardiac remodelling process and cardiomyocyte contraction.16 Dietary components modulate miRNAs levels, inducing either beneficial or deleterious effects in heart function.17,18 Therefore, in the failing human or mouse heart, some miRNA expression patterns, closely related to inflammatory signalling, may result altered in response to high fat feeding.19 Several investigators have confirmed that various miRNAs control the post-transcriptional P2X7R expression.20,21 On the other hand, an interesting report explained that, in a specific inflammatory pathological milieu, a cross-regulation between P2X7R expression and miRNAs may occur;22 such interplay could be potentially relevant also in the cardiovascular system.
Despite some studies have attempted to highlight the relationship between P2X7R activation, the resulting pro-inflammatory cascade, the related miRNAs expression, and the development of cardiac dysfunction, information on a putative effect of such platform on in vivo myocardial performance are scanty, and the role of P2X7R in the heart of animals carrying a diet-induced systemic inflammation still deserves elucidation. The aim of the present study was, therefore, to evaluate the involvement of P2X7R as a cardiac function regulator, in the context of the systemic inflammation induced by high fat diet.
Materials and Methods
Animal Model
Six-weeks-old wild-type (WT) C57BL/6J (ENVIGO, Udine, Italy) and P2X7R knockout (KO) (Jackson Laboratory, Bar Harbor, ME, USA) were utilized. Mice, housed in a germfree stabularium in accordance with the Principles of Laboratory Animal Care (NIH Publication number 85–23, revised 2011) and acclimatized for two weeks, were fed with water and food ad libitum for 16 weeks, and randomly divided in at least four groups of six animals each: WT or KO treated with normal-fat diet (NFD) (5.5% of energy as fat, ENVIGO, Udine, Italy) or high fat diet (HFD) (60% of energy as fat, PF4215, Mucedola, Settimo Milanese, Italy), respectively. Body weight and food intake were monitored at time 0 and weekly. The group assignment was blinded to the operators carrying morpho-functional and molecular analyses. The study protocol was approved by the Ethics Committee of the University of Pisa (1474/2016); we also followed all the international and national guidelines for the care and use of animals (Italian Ministry for Animal Care #943/2015-PR).
Ultra High-Frequency Ultrasound (UHFUS) Examination
All animals were subjected to a cardiac UHFUS examination with a high-resolution imaging system (Vevo 2100, FUJIFILM VisualSonics Inc., Toronto, Canada), as previously reported.23 In detail, trans-mitral inflow pulsed wave Doppler (PW-Doppler) in apical 4-chamber view was used for the evaluation of the left ventricular (LV) diastolic function; the ratio of the early to late ventricular filling velocities (E/A) was calculated from mitral inflow data and used as a diastolic function parameter. Heart B-mode images were acquired with a 55 MHz UHFUS probe (MS550, FUJIFILM VisualSonics) in parasternal long axis (PLAX) and short axis (SAX) views and were analysed offline to calculate the left ventricular mass (LVmass) and the following systolic function parameters: cardiac output (CO), fractional shortening (FS), fractional area change (FAC), stroke volume (SV), and ejection fraction (EF).24 These parameters, in analogy with their use in humans, provide an overall LV morpho-functional outlook. Precisely, they allow to evaluate – in vivo and in near-physiological conditions – the presence of any alterations of the LV, both during diastole and systole. Along with their high sensitivity, they are ideal parameters for comparing groups of animals subjected to treatments that can induce alterations in LV structure and/or function. At the end of treatment, mice were anaesthetized with isoflurane and hearts were removed.
Biochemistry
Blood samples were obtained the day after the ultrasonographic evaluation, to avoid pre-test stress and were collected in K2-EDTA tubes from the tail vein after four hours fasting. Quantitative determinations of glucose (GLUC), cholesterol (CHL) and triglycerides (TRG) were performed by a digital multimeter (multiCare-in, Biochemical Systems International, Arezzo, Italy). Aspartate aminotransferase (AST) activity was measured by an enzymatic rate method in a Synchron Clinical System (Beckman Coulter, Milano, Italy).
Quantitative Real-Time PCR
Total RNA was extracted from the mouse heart using the standard TRIzol method (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Afterwards, the purified RNA (500ng) was reverse transcribed in a 20μL reaction tube using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA), through random priming. Expression of each specific target gene was evaluated by quantitative PCR on Eco real-time instrument (Illumina Inc., San Diego, CA, USA), according to the following protocol: 4µL of 1:5 diluted cDNA template were mixed with 10µL of 2X SSO advanced universal probe supermix (Bio-Rad, Hercules, CA, USA), 1µL of 20X primer and probe mixture (pre-developed TaqMan® gene expression assays, Applied Biosystems), to a final volume of 20µL. Thermal cycling conditions used were 95°C for 30s, followed by 40 cycles at 95°C for 15s and 60°C for 1min. Each sample was analysed in triplicate, and relative gene expression was calculated using the 2−ΔΔCt method and reported as arbitrary units (A.U.). The housekeeping gene β-actin was employed for normalization. Supplementary Table 1 reports the used TaqMan® assays.
Oil Red Staining
To estimate the cardiac lipid content, 12-μm-thick frozen tissue sections were equilibrated and fixed with 4% formaldehyde for 1min at RT. After washing three times with water, the sections were treated with 60% isopropanol solution for 1min, completely covered by 1% filtered Oil red O solution (Bio-Optica, Milan, Italy) and incubated for 6min at RT. Then, the tissue slides were immediately unstained with 60% isopropanol solution for 1min and gently rinsed under running tap water.25 Red stained lipids were examined with light microscopy and quantified by image-dedicated software (ImageJ v1.49 software). Cardiac tissue slides of db/db mice (ENVIGO) were used as positive control.
Picrosirius Red Staining
To estimate the cardiac collagen content, 12-μm-thick frozen tissue sections were treated as paraffin embedded sections: to remove background staining, after 10min step in xylene, sections were hydrated through a descending ethanol series, washed in distilled water, and fixed with 4% formaldehyde at RT for 10min. Subsequently, the 0.1% sirius red picrate solution (Bio-Optica) was applied and allowed to act for 60min. Sections were then washed for 2min in 0.01 N HCl, dehydrated, cleared in xylene for 5min at RT and finally covered with BioMount mounting medium (Bio-Optica) and a glass cover slip.25 For each sample, six randomly selected fields were analysed using a light microscope with a x40 objective lens; blinded measurements were performed. Cardiac tissue slides of db/db (ENVIGO) mice were used as positive control.
Quantification of Tissue Staining
Using an automated action in Photoshop software, each RGB image was transformed into a grey scale image, representative of red pixel only. To quantify the tissue staining, a threshold area was established. Briefly, for each image, the grey value in three different non-signal area was measured, and the average value was used to set the threshold; in the binary image obtained, the percentage area of positive pixel was calculated. This methodology is better detailed elsewhere.26 All image analyses were performed using ImageJ v1.49 software.
Cardiac miRNAs Expression
Cardiac miRNAs were isolated by miRNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instruction. Quantitation of miRNAs was performed using TaqMan® MicroRNA Assays (Applied Biosystems), which included cDNA template preparation by TaqMan® Advanced miRNA cDNA Synthesis kit (Applied Biosystems) and subsequent real-time PCR reaction. In detail, 5µL of 1:10 diluted cDNA template, 10µL of 2X TaqMan® Universal master mix (Applied Biosystems) and 1µL of 20X miRNA TaqMan® probe (Applied Biosystems) were mixed to a final volume of 20µL and incubated at 95°C for 10min for polymerase activation, followed by for 40 cycles at 95°C for 15s and 60°C for 1min. The TaqMan® miRNA assays are reported in Supplementary Table 1. miRNA levels were expressed employing the 2−ΔΔCt method and reported as arbitrary units (A.U.). Two reference miRNAs (miR-16-5p and miR-191-5p) were selected based on the scientific literature,27 after assessment of low variability in our samples.
Immunoblotting and Immunofluorescence
Slides of mouse LV tissue were homogenized in lysis buffer (50mM TRIS-HCl pH 7.4, 250mM NaCl, 1% NP-40) containing protease and phosphatase inhibitor mixture (Sigma-Aldrich Merck, Darmstadt, Germany) using TissueLyser LT (Qiagen, Hilden, Germany). Resulting protein extracts (30 μg/lane) were separated under denaturing conditions on 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore, Bedford, MA, USA). After blocking with BSA 5% in T-tris buffered saline (TBS and Tween-20 0.1%) for 1h at RT, blots were incubated overnight at 4°C with one of the following primary antibodies diluted 1:1000: anti-IL-1β (number 12242), anti-IL-6 (number 12912), and anti-p38 mitogen-activated protein kinase (p38MAPK) (number 9212), anti-p-p38MAPK (number 9211), anti-transforming growth factor-β (TGFβ) (number 3711), anti-monocyte chemoattractant protein-1 (MCP-1) (number 2029) from Cell Signaling Technology (Danvers, MA, USA); anti-NLRP3 (AG-20B-0014) from Adipogen (San Diego, CA, USA); anti-nitric oxide synthase-2 (NOS-2) (sc-651) from Santa Cruz Biotechnology (Dallas, TX, USA), and anti-tumor necrosis factor-α (TNFα) (PA5-19810) from ThermoFisher Scientific. The membranes were then rinsed three times for 10min with T-TBS and incubated with the appropriate HRP-conjugated secondary antibody diluted 1:5000 for 1h at RT. Immune-reactive protein was detected using an enhanced chemiluminescence detection system (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were then incubated at 70°C for 10min in a stripping buffer (5mM Tris–HCl (pH 6.8), 2% SDS, 0.8% β-mercaptoethanol) and re-tested for internal loading control glyceraldehyde 3-phosphate dehydrogenase (GAPDH, antibody number MAB374, Millipore). Films were scanned and band intensity was evaluated using ImageJ v1.49 software. Each sample value was normalised by dividing the intensity of the band of interest with the intensity of GAPDH, or Ponceau S staining. Data were expressed as arbitrary units (A.U.).
For the P2X7R immunofluorescence assay, slides of mouse LV tissue were fixed in 4% paraformaldehyde for 20min at RT, washed with PBS, blocked with BSA 1% in PBS and then incubated overnight with 1:100 rabbit primary antibody (APR-004, Alomone Labs, Jerusalem), followed by 1:500 goat anti-rabbit Alexa Fluor 594 secondary dye antibody for 1h at RT. 4,6-diamidino-2-phenylindole (DAPI) (ThermoFisher Scientific) was used as nuclear marker. Samples were directly observed by fluorescence microscopy using a ×40 objective (EVOS M5000, ThermoFisher Scientific).
Statistical Analysis
After evaluation of normality using Shapiro–Wilk test, statistical differences among the groups were measured using two-way analysis of variance (ANOVA), with genotype and diet as sources of variation, followed by Tukey’s post hoc test. Linear correlations were tested by Pearson’s or Spearman correlation test. A P value ≤0.05 was considered statistically significant. Statistical analyses were performed using Prism software version 7.0 (GraphPad, San Diego, CA, USA).
Results
Metabolic Profile
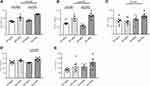
Murine phenotype is shown in Figure 1. At time 0, WT animals weighted 29.2±2.0 g and KO 29.2±1.6 g. At the end of the treatment, WT-HFD mice showed significant weight gain (p < 0.0005) and higher glucose levels (p < 0.002) as compared with WT-NFD mice (Figure 1A and B). In KO-HFD mice, the increase in weight was even more evident (50.2±0.4 g in KO-HFD vs 41.5±2.3 g in WT-HFD, p < 0.002); blood glucose was further increased and circulating triglycerides were considerably higher than those of WT-HFD (Figure 1B and C, all p < 0.05). HFD induced a rise in cholesterol levels only in KO mice (Figure 1D). This altered metabolic state was not associated with increased lipid deposition in cardiac tissue, as revealed by oil red staining (Supplementary Figure 1). No significant changes in aspartate transaminase levels were observed among the four groups of animals (Figure 1E). In summary, KO-HFD mice show higher glucose and triglycerides levels, and gain more weight, than WT-HFD mice.
Heart Performance
Figure 2 shows parameters of heart function as measured in vivo before animal sacrifice. KO-HFD showed a clear alteration of the diastolic pattern compared to WT-HFD animals, as demonstrated by the significant increase in the E/A ratio (1.99±0.37 in KO-HFD vs 1.36±0.05 in WT-HFD, p < 0.03) (Figure 2A). This was associated with a marked morphological remodelling of the myocardium, as indicated by the significant increase of LV mass (170.9±6.4 mg in KO-HFD vs 139.8±3.4 mg in WT-HFD, p < 0.05) (Figure 2B). All the other parameters of systolic function resulted similarly in the four groups of animals (Figure 2C–G). In summary, KO-HFD mice show increased LV mass and alterations of the diastolic pattern.
Inflammatory Profile
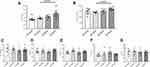
Expression profile of NLRP3-inflammasome complex, regulated by P2X7R was examined. Data are shown in Figure 3. NLRP3 and Caspase-1 gene expression was higher in WT-HFD than in WT-NFD animals (Figure 3A and B, both p < 0.05), and it was associated to a tendentially increased expression of priming signal factor NFκB (Figure 3C).
We also confirmed an increased P2X7R and NLRP3 protein expression in these WT-HFD animals (Supplementary Figure 2).
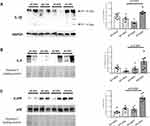
In contrast, heart mRNA of NLRP3, Caspase-1 and NFκB was strongly downregulated in KO-HFD mice, highlighting a key role of P2X7R in the modulation of cardiac inflammasome activation (Figure 3A–C). Interestingly, the transcriptional level of cardiac pro-inflammatory cytokines IL-1β and IL-6 was higher in KO-HFD than in WT-HFD mice (both p < 0.05) (Figure 3D and E); a trend was observed for IL-18, too (Figure 3F). As reported in Figure 4, such increased gene expression translated into a similarly increased IL-1β and IL-6 protein expression at the heart level (Figure 4A and B). To better detail the intracellular signalling mediating such increased protein expression, we found that the p38 pathway was upregulated in KO-HFD animals (Figure 4C).
The following step was to investigate the expression profile of key regulators of cardiac inflammatory pathways involved in the modulation of oxidative stress, endothelial function and chemotactic processes. Results are shown in Figure 5. Compared to control, TNFα (p < 0.02), NOS-2 (p < 0.02) and MCP-1 (p < 0.05) genes were significantly more expressed in the heart of WT-HFD mice (Figure 5A–C). On the other hand, in P2X7R KO animals, we did not observe any increase in TNFα and NOS-2, whereas MCP-1 expression was slightly lower than that of WT-HFD mice (Figure 5A–C). The cardiac NOS-3, ADGRE1 and ICAM-1 levels were unaffected by HFD in either WT or KO animals (Figure 5D–F).
We also tested protein expression of the differentially expressed genes (TNFα, NOS-2 and MCP-1); data are shown in Figure 6 panels A-C and confirm the expression of the relative genes.
In summary, KO-HFD mice show reduced heart expression of NLRP3 and caspase-1 vs WT-HFD; however, this translates into a different mRNA heart content only for TNFα and NOS-2.
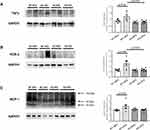
In Figure 7, the transcriptional evaluation of critical fibrogenic mediators involved in the development of myocardial dysfunction is reported. HFD treatment induced a significant increase of heart TGFβ expression in WT mice (p < 0.01), but no effects were observed in KO animals (Figure 7A). The evaluation of the downstream pro-fibrotic LOXL2/collagen/MMP-9 signalling pathway showed as HFD induced a small, albeit significant, increment of LOXL2 and collagen 1 in KO animals (Figure 7B–D). MMP-9 did not significantly vary (Figure 7E).
In Figure 8, we reported an evaluation of the degree of fibrosis. Picrosirius shows an increased collagen deposition in WT-HFD animals (Figure 8A), coupled with an increased TGFβ protein expression (Figure 8B) (p < 0.01); such HFD-induced damage was not evident in HFD-P2X7 KO animals. In summary, KO-HFD animals do not display an increased heart fibrosis, except for a small rise of LOXL2 and collagen 1.
Heart miRNAs Expression
Lastly, we evaluated the expression profile of five specific miRNAs involved in the pathogenesis of cardiovascular damage, closely associated to tissue process of fibrosis and hypertrophy, and related to NLRP3 inflammasome regulation.28,29 As reported in Figure 9, after treatment with HFD, a clear increase of miR-27a and miR-27b were detected in the heart of WT mice (Figure 9A and B); the same trend was observed for miR-214 (Figure 9C). Conversely, miR-126 expression was significantly reduced compared to control animals (Figure 9D), whereas the expression of miR-21 was unchanged (Figure 9E). In KO-HFD mice, the cardiac expression level of miR-27a and miR-27b appeared comparable to that observed in WT-HFD mice (Figure 9A and B); the expression of miR-214 and miR-126 was intensified and further attenuated, respectively, compared to WT-HFD mice (Figure 9C and D). In summary, KO-HFD show increased miR-214 and reduced miR-126 vs WT-HFD mice.
Regression Analysis
The potential relationships among impaired myocardial performance, altered metabolic parameters, increased inflammatory mRNA cytokine levels and dysregulated miRNA expression were detected by Pearson’s correlation test. Notably, in KO-HFD mice, LV mass positively correlated with weight (r = 0.83, p < 0.05), GLUC (r = 0.76), TRG (r = 0.75) (Figure 10A); IL-1β (r = 0.88, p < 0.05), IL-6 (r = 0.75) and IL-18 (r = 0.77) (Figure 10B). On the other hand, we found that, in KO-HFD mice, the downregulated miR-126 expression was significantly correlated with cardiac anatomic changes (r = −0.75 for LV mass, p < 0.05) and, although to a lesser extent, with a diastolic dysfunction (r = −0.65 for E/A ratio) (Figure 10C).
Discussion
The present study points out the key role of P2X7R in protecting the heart toward some aspects of the inflammatory process triggered by a high-fat diet. The main results can be summarized as follows: an excess of dietary fat per se is unable to induce myocardial dysfunction in WT mice, while P2X7R KO mice develop an impairment of diastolic function and an increased left ventricular mass; such functional alterations involve the downregulation of the NLRP3-Caspase-1 inflammasome. Intriguingly, such cascade is not responsible for heart expression of IL-1β and IL-6, still upregulated in this animal model. The heart of P2X7R KO animals neither shows any rise in TNFα, NOS-2 and MCP-1, suggesting a P2X7R active involvement also in the activation of such inflammatory-oxidative pathways, barely explored so far at the heart level.30 On the other hand, high fat diet does not increase the heart expression of TGFβ, neither induce any overt heart fibrosis in these animals.
Comparing the dysfunctional parameters registered at the heart level in these mice before sacrifice, it appears that the P2X7R-mediated activation of the inflammatory-oxidative process, is not sufficient, by itself, to induce a cardiac functional alteration after high-fat diet. Previous studies on this topic have generated conflicting results. P2X7R has been mainly recognized, in several tissues and organs including the heart, as a pro-inflammatory receptor,11,31,32 and in a model of myocardial ischemia it has been related to the activation of the sympathetic system.33,34 The increased heart expression of P2X7R, NLRP3 and caspase-1 has been already described in rats;35 however, in that study, cardiac ultrasonography was not performed; moreover, mice is the only available animal P2X7R KO model. Surprisingly, in the heart of P2X7R KO, the high-fat diet induced, obviously via alternative pathways other than P2X7R, a high IL-1β and IL-6 gene and protein expression. Several studies have pointed out the role of some pro-inflammatory cytokines in the pathogenesis of cardiomyopathies: in different murine models, acute and chronic administration of IL-1β determined a reduced ventricular contractility,36 while systolic and diastolic function improved after blocking IL-1β signal by genetic deletion or pharmacologic inhibition.37 In experimental models of dysfunctional heart, high IL-6 levels were associated with altered ventricular remodelling, increased stiffness, and enhanced hypertrophic response.38 Our results suggest that, in vivo, the presence of P2X7R might play a sort of cardioprotective role, as already observed in other experimental settings; for example, in response to ischaemia-reperfusion-induced myocardial damage.39 The clear alteration of diastolic pattern (significantly increased E/A ratio) and an evident morphologic remodelling (as from increased LV mass) observed only in P2X7R KO animals suggests that other pathways might contribute to the development of such morphological and functional alterations; the increased IL-6 and IL-1β content in the heart of these KO animals reinforces this hypothesis; less likely, the presence of the P2X7R should be regarded as necessary for avoiding such condition.
To gain further insight the putative molecular post-transcriptional regulation of these cytokines, we measured tissue expression of p38 MAPK, key component of the transduction pathway implied in the cytokine-mediated inflammation;40 such kinase was significantly more expressed and more activated in the heart of KO-HFD animals, reinforcing the hypothesis of its involvement as determinant of diastolic dysfunction.41
The role of P2X7R in the pathogenesis of tissue fibrosis is raising attention;42 therefore, we also measured the degree of fibrosis induced by high fat diet. Despite the early functional alterations observed in vivo, these KO animals seem to be protected by the extreme consequences of oxidative stress in myocardium. In fact, they show a small rise in some markers of fibrosis (collagen 1 and LOXL2), but the main profibrotic factor, ie, TGFβ, is not upregulated in these KO-HFD animals, likely contributing to limit the degree of myocardial fibrosis.
Interestingly, inflammatory cytokines may also interact with miRNAs in exerting their detrimental action on the heart;43 searching for a putative regulatory functional relationship, we measured some cardiac-specific miRNAs, relating them to IL-1β and IL-6 tissue expression. We found an upregulation of miR-214 and a down-regulation of miR-126 in KO vs WT mice treated with high fat diet. The relationship between miR-214, miR-126 and inflammatory cytokines has been reported in other CV diseases; recently, Zheng et al44 have shown as miR-214 was hyper-expressed in interstitial cells of aortic valve, inducing an excess of IL-6-mediated inflammatory response and promoting valvular calcification and progression of calcific aortic valve disease. A miR-214 up-regulation enhanced myocardial damage related to viral myocarditis by stimulating IL-1β and IL-6 expression.45 On the other hand, miR-126 negatively modulates the effect of an excess of glucose on the production of inflammatory markers, including IL-6,46 and exerts a protective role toward hypoxia-induced myocardial damage, reducing the cytokines-mediated inflammation.47
The metabolic phenotype of these animals might help in explaining our results. In fact, at the end of the treatment with high-fat diet, P2X7R KO animals showed a worse phenotype than WT littermates. It should be pointed out as the P2X7R KO mouse model, even when fed with normal chow, displays a phenotype characterized by a slightly higher weight, lower cholesterol levels and a specific adipose tissue distribution with respect to WT littermate.48 We also tested for an excess of lipotoxicity in the heart, but this was not found. Such picture emphasizes the importance of systemic metabolic dysregulation in the development of cardiac functional alterations.5,49 We might hypothesize that, in the context of marked systemic metabolic derangement, heart tissue homeostasis is altered by the lack of the P2X7R; such condition might deregulate miR-126 and miR-214; as consequence, IL-1β and IL-6 expression would be enhanced through p38 MAPK, explaining, at least in part, the onset of heart dysfunction. Of note, such link between miR-126, pro-inflammatory cytokines and p38 MAPK has been suggested as a mechanism concurring to the progression of atherosclerosis.50
We should acknowledge some limitations of the present study: first, our observations were obtained using a reduced sample size that, by the way, does not allow to detect potential significant differences between some parameters analysed. Second, our experimental design does not include any treatment with inhibitors or activators of molecular pathways involved in cardiac regulation, to evaluate the resulting functional effects. Third, this murine model does not allow more prolonged observations in aging animals. Finally, the results obtained in the present study, using a murine model, cannot be automatically extended to the human condition.
Conclusions
P2X7R seems to protect the murine heart toward some aspects of the inflammatory damage induced by a high fat diet. This observation opens a novel prospective on the putative role played by P2X7R, ubiquitous receptor expressed in almost all tissues and organs, in maintaining tissue homeostasis and preserved organ functionality, looking at the numerous P2X7R agonist and antagonists under development as drivers of relevant and multifaceted future clinical implications.
Funding
This research has been supported by a grant from the University of Pisa (fondi Ateneo 2018; recipient AS).
Disclosure
The authors report no conflict of interest in this work.
References
1. Nishida K, Otsu K. Inflammation and metabolic cardiomyopathy. Cardiovasc Res. 2017;113:389–398. doi:10.1093/cvr/cvx012
2. Luc K, Schramm-Luc A, Guzik TJ, Mikolajczyk TP. Oxidative stress and inflammatory markers in prediabetes and diabetes. J Physiol Pharmacol. 2019;70:809–824.
3. Crewe C, An YA, Scherer PE. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J Clin Invest. 2017;127:74–82. doi:10.1172/JCI88883
4. Singh M, Benencia F. Inflammatory processes in obesity: focus on endothelial dysfunction and the role of adipokines as inflammatory mediators. Int Rev Immunol. 2019;38:157–171. doi:10.1080/08830185.2019.1638921
5. Chen M, Li Y, Sun Q, et al. Dairy fat and risk of cardiovascular disease in 3 cohorts of US adults. Am J Clin Nutr. 2016;104:1209–1217. doi:10.3945/ajcn.116.134460
6. Liu XM, Liu YJ, Huang Y, et al. Dietary total flavonoids intake and risk of mortality from all causes and cardiovascular disease in the general population: a systematic review and meta-analysis of cohort studies. Mol Nutr Food Res. 2017;61(6):1601003. doi:10.1002/mnfr.201601003
7. Gonçalves N, Silva AF, Rodrigues PG, et al. Early cardiac changes induced by a hypercaloric Western-type diet in “subclinical” obesity. Am J Physiol Heart Circ Physiol. 2016;310:H655–H666. doi:10.1152/ajpheart.00684.2015
8. Carbone S, Canada JM, Buckley LF, et al. Dietary fat, sugar consumption, and cardiorespiratory fitness in patients with heart failure with preserved ejection fraction. JACC Basic Transl Sci. 2017;2(1):30513–30525.
9. Poret JM, Souza-Smith F, Marcell SJ, et al. Primeaux, high fat diet consumption differentially affects adipose tissue inflammation and adipocyte size in obesity-prone and obesity-resistant rats. Int J Obes. 2018;42:535–541. doi:10.1038/ijo.2017.280
10. Avtanski D, Pavlov VA, Tracey KJ, Poretsky L. Characterization of inflammation and insulin resistance in high-fat diet-induced male C57BL/6J mouse model of obesity. Animal Model Exp Med. 2019;25:252–258. doi:10.1002/ame2.12084
11. Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity. 2017;47:15–31. doi:10.1016/j.immuni.2017.06.020
12. Solini A, Menini S, Rossi C, et al. The purinergic 2X7 receptor participates in renal inflammation and injury induced by high-fat diet: possible role of NLRP3 inflammasome activation. J Pathol. 2013;231:342–353. doi:10.1002/path.4237
13. Ward JR, West PW, Ariaans MP, et al. Temporal interleukin-1beta secretion from primary human peripheral blood monocytes by P2X7-independent and P2X7-dependent mechanisms. J Biol Chem. 2010;23:23147–23158. doi:10.1074/jbc.M109.072793
14. Zhang X, Fu Y, Li H, et al. H3 relaxin inhibits the collagen synthesis via ROS- and P2X7R-mediated NLRP3 inflammasome activation in cardiac fibroblasts under high glucose. J Cell Mol Med. 2018;22:1816–1825. doi:10.1111/jcmm.13464
15. Mezzaroma E, Toldo S, Farkas D, et al. The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc Natl Acad Sci USA. 2011;108:19725–19730. doi:10.1073/pnas.1108586108
16. Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116:751–762. doi:10.1161/CIRCRESAHA.116.303549
17. Zheng Z, Ge Y, Zhang J, et al. PUFA diets alter the microRNA expression profiles in an inflammation rat model. Mol Med Rep. 2015;11:4149–4157. doi:10.3892/mmr.2015.3318
18. Alehagen U, Johansson P, Aaseth J, Alexander J, Wågsäter D. Significant changes in circulating microRNA by dietary supplementation of selenium and coenzyme Q10 in healthy elderly males. A subgroup analysis of a prospective randomized double-blind placebo-controlled trial among elderly Swedish citizens. PLoS One. 2017;12:1–18.
19. Guedes EC, França GS, Lino CA, et al. MicroRNA expression signature is altered in the cardiac remodeling induced by high fat diets. J Cell Physiol. 2016;231:1771–1783. doi:10.1002/jcp.25280
20. Rahman OA, Sasvari-Szekely M, Szekely A, Faludi G, Guttman A, Nemoda Z. Analysis of a polymorphic microRNA target site in the purinergic receptor P2RX7 gene. Electrophoresis. 2010;31:1790–1795. doi:10.1002/elps.200900664
21. Huang S, Chen Y, Wu W, et al. miR-150 promotes human breast cancer growth and malignant behavior by targeting the pro-apoptotic purinergic P2X7 receptor. PLoS One. 2013;8:e80707. doi:10.1371/journal.pone.0080707
22. Killeen ME, Ferris L, Kupetsky EA, Falo LJ, Mathers AR. Signaling through purinergic receptors for ATP induces human cutaneous innate and adaptive Th17 responses: implications in the pathogenesis of psoriasis. J Immunol. 2013;190:4324–4336. doi:10.4049/jimmunol.1202045
23. Faita F, Di Lascio N, Rossi C, Kusmic C, Solini A. Ultrasonographic characterization of the db/db mouse: an animal model of metabolic abnormalities. J Diabetes Res. 2018;8:4561309.
24. Hein SJ, Lehmann LH, Kossack M, et al. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS One. 2015;10:e0122665. doi:10.1371/journal.pone.0122665
25. Wadsworth MP, Sobel BE, Schneider DJ, Tra W, Hirtum HV, Taatjes DJ. Quantitative analysis of atherosclerotic lesion composition in mice. In: Cell Imaging Techniques. Humana Press; 2006:137–152.
26. Solini A, Rossi C, Duranti E, Taddei S, Natali A, Virdis A. Saxagliptin prevents vascular remodeling and oxidative stress in db/db mice. Role of endothelial nitric oxide synthase uncoupling and cyclooxygenase. Vascul Pharmacol. 2016;76:62–71. doi:10.1016/j.vph.2015.10.002
27. Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi:10.1261/rna.939908
28. Ono K, Kuwabara Y, Han J. MicroRNA and cardiovascular diseases. FEBS J. 2011;278:1619–1633. doi:10.1111/j.1742-4658.2011.08090.x
29. Liu D, Zeng X, Li X, et al. Advances in the molecular mechanisms of NLRP3 inflammasome activators and inactivators. Biochem Pharmacol. 2020;175:113863. doi:10.1016/j.bcp.2020.113863
30. Cheng W, Sun Y, Wu Q, et al. Paraventricular nucleus P2X7 receptors aggravate acute myocardial infarction injury via ROS-induced vasopressin-V1b activation in rats. Neurosci Bull. 2021;37:641–656. doi:10.1007/s12264-021-00641-8
31. Zhou J, Tian G, Quan Y, et al. Inhibition of P2X7 purinergic receptor ameliorates cardiac fibrosis by suppressing NLRP3/IL-1β pathway. Oxid Med Cell Longev. 2020;2020:7956274. doi:10.1155/2020/7956274
32. Zhou J, Zhou Z, Liu X, Yin HY, Tang Y, Cao X. P2X7 receptor-mediated inflammation in cardiovascular disease. Front Pharmacol. 2021;12:654425. doi:10.3389/fphar.2021.654425
33. Tu G, Li G, Peng H, et al. P2X(7) inhibition in stellate ganglia prevents the increased sympathoexcitatory reflex via sensory-sympathetic coupling induced by myocardial ischemic injury. Brain Res Bull. 2013;96:71–85. doi:10.1016/j.brainresbull.2013.05.004
34. Liu J, Li G, Peng H, et al. Sensory-sympathetic coupling in superior cervical ganglia after myocardial ischemic injury facilitates sympathoexcitatory action via P2X7 receptor. Purinergic Signal. 2013;9:463–479. doi:10.1007/s11302-013-9367-2
35. Chen X, Li H, Wang K, et al. Aerobic exercise ameliorates myocardial inflammation, fibrosis and apoptosis in high-fat-diet rats by inhibiting P2X7 purinergic receptors. Front Physiol. 2019;10:1286. doi:10.3389/fphys.2019.01286
36. Fernández-Sada E, Torres-Quintanilla A, Silva-Platas C, et al. Proinflammatory cytokines are soluble mediators linked with ventricular arrhythmias and contractile dysfunction in a rat model of metabolic syndrome. Oxid Med Cell Longev. 2017;2017:7682569. doi:10.1155/2017/7682569
37. Toldo S, Mezzaroma E, Bressi E, et al. Interleukin-1β blockade improves left ventricular systolic/diastolic function and restores contractility reserve in severe ischemic cardiomyopathy in the mouse. J Cardiovasc Pharmacol. 2014;64:1–6. doi:10.1097/FJC.0000000000000106
38. Meléndez GC, McLarty JL, Levick SP, Du Y, Janicki JS, Brower GL. Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension. 2010;56:225–231. doi:10.1161/HYPERTENSIONAHA.109.148635
39. Vessey DA, Li L, Kelley M. P2X7 receptor agonists pre- and postcondition the heart against ischemia-reperfusion injury by opening pannexin-1/P2X7 channels. Am J Physiol Heart Circ Physiol. 2011;301:H881–H887. doi:10.1152/ajpheart.00305.2011
40. Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi:10.1038/sj.cr.7290257
41. Liu ZF, Ji JJ, Zheng D, Su L, Peng T. Calpain-2 protects against heat stress-induced cardiomyocyte apoptosis and heart dysfunction by blocking p38 mitogen-activated protein kinase activation. J Cell Physiol. 2019;234:10761–10770. doi:10.1002/jcp.27750
42. Gentile D, Natale M, Lazzerini PE, Capecchi PL, Laghi-Pasini F. The role of P2X7 receptors in tissue fibrosis: a brief review. Purinergic Signal. 2015;11:435–440. doi:10.1007/s11302-015-9466-3
43. Gorabi AM, Kiaiem N, Sathyapalan T, Al-Rasadi K, Jamialahmadi T, Sahebkar A. The role of MicroRNAs in regulating cytokines and growth factors in coronary artery disease: the ins and outs. J Immunol Res. 2020;2020:5193036. doi:10.1155/2020/5193036
44. Zheng D, Zang Y, Xu H, et al. MicroRNA-214 promotes the calcification of human aortic valve interstitial cells through the acceleration of inflammatory reactions with activated MyD88/NF-κB signaling. Clin Res Cardiol. 2019;108:691–702. doi:10.1007/s00392-018-1398-9
45. Chen ZG, Liu H, Zhang JB, Zhang SL, Zhao LH, Liang WQ. Upregulated microRNA-214 enhances cardiac injury by targeting ITCH during coxsackievirus infection. Mol Med Rep. 2015;12:1258–1264. doi:10.3892/mmr.2015.3539
46. Li Y, Zhou Q, Zhe Pei C, et al. Hyperglycemia and advanced glycation end products regulate miR-126 expression in endothelial progenitor cells. J Vasc Res. 2016;53:94–104. doi:10.1159/000448713
47. Luo Q, Guo D, Liu G, Chen G, Hang M, Jin M. Exosomes from MiR-126-overexpressing adscs are therapeutic in relieving acute myocardial ischaemic injury. Cell Physiol Biochem. 2017;44:2105–2116. doi:10.1159/000485949
48. Beaucage KL, Xiao A, Pollmann SI, et al. Loss of P2X7 nucleotide receptor function leads to abnormal fat distribution in mice. Purinergic Signal. 2014;10:291–304. doi:10.1007/s11302-013-9388-x
49. Rayner JJ, Banerjee R, Holloway CJ, et al. The relative contribution of metabolic and structural abnormalities to diastolic dysfunction in obesity. Int J Obes. 2018;42:441–447. doi:10.1038/ijo.2017.239
50. Hao X-Z, Fan H-M. Identification of miRNAs as atherosclerosis biomarkers and functional role of miR-126 in atherosclerosis progression through MAPK signalling pathway. Eur Rev Med Pharmacol Sci. 2017;21:2725–2733.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.