Back to Journals » Infection and Drug Resistance » Volume 11
Overview of the development of quinolone resistance in Salmonella species in China, 2005–2016
Authors Song Q, Xu Z, Gao H, Zhang D
Received 19 November 2017
Accepted for publication 22 December 2017
Published 26 February 2018 Volume 2018:11 Pages 267—274
DOI https://doi.org/10.2147/IDR.S157460
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Qifa Song,1 Zhaojun Xu,2 Hong Gao,1 Danyang Zhang1
1Department of Microbiology, Ningbo Municipal Centre for Disease Control and Prevention, Ningbo, Zhejiang, China; 2Intensive Care Unit, Ningbo No. 2 Hospital, Zhejiang, China
Purpose: Several factors contribute to the complexity of quinolone resistance in Salmonella, including >2000 different Salmonella serotypes, a variety of hosts for Salmonella, and wide use of quinolones in human beings and animals. We thus aimed to obtain an overview of the development of quinolone resistance and relevant molecular mechanisms of such a resistance in Salmonella species.
Materials and methods: A total of 1,776 Salmonella isolates were collected in Ningbo, China, between 2005 and 2016. Antimicrobial susceptibility to quinolone and relevant genetic mechanisms in these isolates were retrospectively analyzed.
Results: The ratio for ciprofloxacin (CIP) resistant:reduced CIP susceptible:CIP susceptible was 26:522:1,228. CIP resistance was found in nine of 51 serotypes: Derby, London, Kentucky, Indiana, Corvallis, Rissen, Hadar, Typhimurium, and Agona. Of 26 CIP-resistant isolates, all were concurrently resistant to ampicillin and 21 were also concurrently resistant to cefotaxime and produced extended-spectrum β-lactamase (ESBL). The minimal inhibitory concentration values were at three levels: 2–4 μg/mL (serotypes except for Kentucky and Indiana), 16 μg/mL (one Kentucky isolate), and >32 μg/mL (Indiana isolates). As with the three most common serotypes, Salmonella Typhi showed quickly increased prevalence of reduced CIP susceptibility in recent years, Salmonella Enteritidis remained at a high prevalence of reduced CIP susceptibility throughout the study period, and several isolates of Salmonella Typhimurium were resistant to CIP. Transferable plasmid-mediated quinolone resistance gene qnrB was only found in all CIP-resistant isolates. In contrast, gyrA mutations were often found in reduced CIP-susceptible isolates and were not necessarily found in all CIP-resistant isolates.
Conclusion: We conclude that in Salmonella, there exists a high prevalence of reduced CIP susceptibility and a low prevalence of CIP resistance, which focuses on several serotypes. Our study also demonstrates that, rather than gyrA mutations, qnrB is the most common indicator for CIP resistance.
Keywords: quinolone resistance, Salmonella species, ciprofloxacin-resistant Salmonella isolates, genetic determinant of quinolone resistance
Introduction
Salmonella species (Salmonella spp.) are a group of bacteria that can survive in animals, human beings, and the environment. They can cause from mild to life-threatening salmonellosis, such as enteritis and typhoid fever, both in human beings and animals, and have become a major public health issue worldwide.1 Of >2,000 Salmonella serotypes, <100 serotypes account for most of prevalent and important human infections, including Salmonella Typhi (S. typhi), Salmonella Enteritidis (S. Enteritidis), and Salmonella Typhimurium (S. Typhimurium).2,3
Quinolones are a family of synthetic broad-spectrum antimicrobial drugs.4 The first quinolone is nalidixic acid, which is considered to be the predecessor of all members of the quinolone family. The majority of quinolones in present clinical use are more powerful fluoroquinolones, such as ciprofloxacin (CIP), which remains the first-line drug of choice for treating salmonellosis.5
Quinolone resistance in Salmonella spp. is an increasing problem.6 Based on susceptibility testing results of nalidixic acid and CIP, quinolone resistance can be ranked at three levels: resistant to CIP (resistant to both drugs), of reduced CIP susceptibility (resistant to nalidixic acid and intermediate to CIP), and susceptible to both drugs.7 Recently, particular concern has been raised about highly CIP-resistant strains of several Salmonella serotypes circulating in both human beings and the environment.8,9 To treat salmonellosis cases caused by CIP-resistant Salmonella strains, a third-generation cephalosporin, such as cefotaxime or ceftriaxone, is the first choice.10
CIP resistance in Salmonella is considered to be mainly attributed to point mutations in gyrA, gyrB, and parC in the quinolone resistance-determining regions (QRDRs).11 In addition, transferable plasmid-mediated quinolone resistance (PMQR) genes may cause low-level resistance to fluoroquinolones.11 These genes include pentapeptide repeat protein-encoding qnrA, qnrB, and qnrS; an efflux-pump-encoding qepA; and an aminoglycoside acetyltransferase-encoding enzyme variant aac(6 ¢)-Ib-cr.12,13
A lot of studies have been conducted to explore CIP-resistant Salmonella strains of several particular serotypes. Most of these studies are some kind of “case report analysis” with limited information on overall quinolone resistance in Salmonella species. There are several factors making quinolone resistance complex in Salmonella, including >2,000 different Salmonella serotypes, a variety of hosts for Salmonella, and wide use of quinolones in human beings and animals. This situation calls for a more general conclusion for explaining CIP resistance development in Salmonella species as a whole.
Therefore, through investigating the development of quinolone resistance in all Salmonella species, what serotypes account for most CIP-resistant isolates, and the relevant molecular mechanisms, we aimed to obtain an overview of development of quinolone resistance and the relevant molecular mechanisms in Salmonella.
Materials and methods
Bacterial isolates
A total of 1,776 Salmonella isolates, 1,226 from non-duplicate patient specimens (1,006 feces and 220 blood samples) and 550 from environmental samples (445 food samples and 105 river water samples), were collected in Ningbo, a city in mid-east China that has a population of 9 million and a land area of 9,780 km2, between 2005 and 2016 (Table 1). These isolates were from an enteric disease surveillance project and clinical patients. Approval of clinical isolates from the ethics committee of Ningbo No. 2 Hospital, Zhejiang, China, and written informed consent of patients were obtained. These isolates were identified by API 20E biochemical identification system (bioMerieux, Paris, France) and were typed into 51 serotypes using Salmonella-specific antisera (Denka Seiken, Japan). In Ningbo, the top three most found serotypes were S. typhi (188 isolates), S. Enteritidis (163 isolates), and S. Typhimurium (156 isolates). All S. typhi isolates came from patients. S. Typhimurium and S. Enteritidis isolates came from both patients and environmental sources.
Antimicrobial susceptibility testing (AST)
Since comparative study has proved high categorical agreements between E-test and disk diffusion and broth microdilution for the detection of fluoroquinolone resistance in typhoidal and nontyphoidal Salmonella serotypes,14 to screen for quinolone-resistant isolates, we performed AST on all isolates using the disk diffusion method specified in Clinical and Laboratory Standards Institute (CLSI) document M02-A12.15 AST results were interpreted according to guidelines in CLSI document M100-S27.16 The antibiotic disks (Oxoid, Hampshire, UK) included CIP (5 μg) and nalidixic acid (30 μg), as well as ampicillin (10 μg), cefotaxime (30 μg), cefotaxime–clavulanate (30/10 μg), gentamicin (10 μg), chloramphenicol (30 μg), and trimethoprim–sulfamethoxazole (1.25/23.75 μg). Escherichia coli ATCC 25922 was used as a quality control strain. Isolates with ≥5 mm increase in the zone diameter for cefotaxime–clavulanate vs. the zone diameter for cefotaxime were defined to be producing extended-spectrum β-lactamase (ESBL). Then, minimal inhibitory concentrations (MICs) of CIP and cefotaxime in CIP-resistant isolates were determined using the E-test (Oxoid) method. MICs of nalidixic acid in CIP-resistant isolates were determined using broth microdilution. Based on AST results, isolates were classified into three groups with different CIP resistance levels: susceptibility, reduced CIP susceptibility, and resistance to CIP.
Detection of genetic determinants associated with quinolone and β-lactam resistance
CIP-resistant isolates, 100 randomly selected reduced CIP-susceptible isolates, and 100 randomly selected CIP-susceptible isolates were tested for genes related to quinolone resistance using primers previously described (Table 2), including gyrA, gyrB, and parC; PMQR genes qnrA, qnrB, and qnrS; efflux-pump-encoding qepA; and aminoglycoside acetyltransferase-encoding enzyme variant aac(6 ¢)-Ib-cr. Amplified sequences of gyr and parC were sequenced (Thermo Fisher Scientific, Waltham, MA, USA) and their mutations were determined by aligning against the homologous sequence of S. Typhimurium LT2. Additionally, CIP- and β-lactam-resistant isolates were tested for reported β-lactamase genes (blaCTX-M-like, blaTEM, and blaOXA).
  | Table 2 Primers used for amplifying genes related to quinolone and β-lactam resistance Abbreviations: QRDR, quinolone resistance-determining region; PMQR, plasmid-mediated quinolone resistance. |
Pulsed-field gel electrophoresis (PFGE)
CIP-resistant isolates were subtyped by PFGE using restriction enzyme XbaI following PulseNet standardized protocol.19 Salmonella Braenderup (H9812) was used as a reference strain. The PFGE patterns were determined by BioNumerics software (Applied Maths, Sint-Martens-Latum, Belgium). A dendrogram of PFGE patterns was constructed using the unweighted pair-group method with arithmetic mean (UPGMA) with a position tolerance of 1.2%. PFGE patterns were defined by groups of bands that formed at least 90% Dice similarity cutoffs on the dendrogram. In this study, a cluster was defined as more than one isolate with identical PFGE patterns. Sporadic cases were defined when there was a unique PFGE pattern for one isolate.
Interpretation of results for overall quinolone resistance
We assessed overall quinolone resistance in Salmonella species as follows. First, levels of quinolone resistance in the three most common serotypes, S. typhi, S. Typhimurium, and S. Enteritidis, were derived for different isolation periods. Second, what serotypes accounted for, and what PFGE features were found in, highly quinolone-resistant isolates that were resistant to CIP and nalidixic acid, were assessed. Third, what genes related to quinolone resistance existed in quinolone-resistant isolates.
We used SPSS 20 software to calculate Pearson’s chi-squared test for comparison of resistance rates between different groups of isolates. Statistical significance was accepted at P value of <0.05.
Results
Totally, of 1,776 isolates, 26 (1.5%) were resistant to CIP and 522 (29%) were of reduced CIP susceptibility (Table 3). The three most common Salmonella serotypes were S. typhi, S. Enteritidis, and S. Typhimurium; each had distinct features of quinolone resistance development during three periods from 2005 to 2016 (Table 1). During the three periods of 2005–2008, 2009–2012, and 2013–2016, S. typhi was characteristic of apparent increase in reduced CIP susceptibility: from 9% (5/54) to a moderate level of 35% (21/60) and a high rate of 89% (66/74); S. Enteritidis remained at steady, high rates of reduced CIP susceptibility: 59% (23/39), 56% (41/73), and 67% (34/51); in contrast, S. Typhimurium showed both a high level of reduced CIP susceptibility, i.e., 44% (17/39), 35% (16/46), and 38% (27/71), and a prevalence of 5% of CIP resistance. Overall, CIP resistance was found in nine of 51 serotypes: Derby, London, Kentucky, Indiana, Corvallis, Rissen, Hadar, Typhimurium, and Agona. Especially, six Salmonella Indiana (S. Indiana) isolates were resistant to CIP. The MIC values were at three levels in CIP-resistant isolates: 2–4 μg/mL (seven serotypes except for Kentucky and Indiana), 16 μg/mL (one Kentucky isolate), and >32 μg/mL (all Indiana isolates; Table 4).
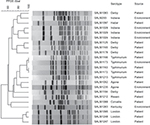
As to other antimicrobial agents, prevalence of resistance (33.8%) to ampicillin was the highest (Table 3). Average prevalence of resistance to other antimicrobials was significantly higher in the group of CIP-resistant isolates than in the other two groups (P<0.01). Of 26 CIP-resistant isolates, all were concurrently resistant to ampicillin and 21 were also concurrently resistant to cefotaxime and produced ESBL. In addition, 19 CIP-resistant isolates were from patients and seven were from environmental sources. All S. Indiana CIP-resistant isolates and a number of S. Typhimurium, S. Agona, and S. Rissen CIP-resistant isolates were from environmental sources (Figure 1).
There were two types of gyrA mutations at loci 83, three types of gyrA mutations at loci 87, and four PMQR genes detected in CIP-resistant and reduced CIP-susceptible isolates (Tables 4 and 5). Most found gyrA mutations were S83F and D87N that could be detected in both groups of isolates. A silent mutation S(TCC)83S(TCT) was found in a number of CIP-resistant Agona and Derby isolates. It is important to note that, most reduced CIP-susceptible S. typhi had S83F or D87Y mutation. Especially, this D87Y mutation was detected only in reduced CIP-susceptible S. typhi. No gyrB mutation was found in such reduced CIP-susceptible S. typhi isolates. Regarding PMQR genes, 100% CIP-resistant isolates harbored qnrB that, in contrast, could not be detected in reduced CIP-susceptible isolates. Gene aac(6 ¢)-Ib-cr in CIP-resistant isolates was also detected at a much higher prevalence in CIP-resistant isolates. qnrS existed in both groups. Especially, in Kentucky and Indiana isolates with CIP MIC of >16 μg/mL, two gyrA mutations, qnrB and usually aac(6 ¢)-Ib-cr, concurrently existed. In isolates of other serotypes with smaller CIP MIC, one or no gyrA mutation and at least qnrB and sometimes plus aac(6´)-Ib-cr existed. No genetic determinants of quinolone resistance were detected in 100 susceptible isolates.
As to β-lactam resistance genes in 26 β-lactam- and CIP-resistant isolates, 21 were detected positive for at least one of TEM, OXA, and CTX-M genes (Table 4).
PFGE results
Although there was no overall clustering in 23 (three failed PFGE) CIP-resistant isolates, PFGE results showed that highly identical PFGE patterns were observed in some common serotypes, such as Typhimurium and Indiana, indicating that CIP-resistant isolates of some serotypes had very limited diversity (Figure 1).
Discussion
We found distinctive features about the overall development of quinolone resistance in Salmonella species: high prevalence of reduced CIP susceptibility and low prevalence of CIP resistance, which focuses on several serotypes, and PMQR gene qnrB being the most common indicator for CIP resistance.
Totally, the ratio for three levels of quinolone susceptibility (CIP resistant:reduced CIP susceptible:CIP susceptible) was 26:522:1,228, indicating a small proportion of CIP-resistant isolates and large proportion of reduced CIP-susceptible isolates. In the literature, a few studies have explored overall CIP resistance development in Salmonella and the limited results are varied. In Hong Kong, a study showed that 42% of 963 non-typhoidal Salmonella isolates are resistant to nalidixic acid (corresponding to reduced CIP susceptibility in our study) and 13.7% are resistant to CIP,6 whereas the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) found that, of 2,236 non-typhoidal Salmonella isolates, only 56 (2%) are resistant to nalidixic acid and eight (0.4%) are resistant to CIP.20 Our results are distinct from the Hong Kong study that shows a high prevalence of both CIP resistance and reduced CIP susceptibility, and the NARMS report that shows a low prevalence of both. A recent study has reported that 16.9% of isolates of animal sources are resistant to CIP, a prevalence much higher than that (1.5%) in our study. This difference suggests a gap in CIP resistance level between isolates from animals and human beings.21
Our results show distinctive phenotypes of quinolone resistance in different serotypes. As to the three most common serotypes, S. typhi showed quickly increased prevalence of reduced CIP susceptibility in recent years, a finding that we have reported in a previous study.22 S. Enteritidis remained at a high prevalence of reduced CIP susceptibility throughout the study period. Nevertheless, in addition to a high prevalence of reduced CIP susceptibility, 5% of S. Typhimurium isolates were resistant to CIP. As early as 2005, up to 70% of 44 S. Typhimurium isolates from outpatients were reported to be resistant to CIP.23 However, our findings illustrate that CIP resistance in S. Typhimurium has emerged only during recent years. In our study, the most serious situation of CIP resistance was observed in S. Indiana, a result consistent with a recent report.24
CIP resistance was found centrally in limited serotypes. PFGE results of CIP-resistant isolates show no overall clustering and a predominant clone in several serotypes, as indicated by limited diversity among multiple isolates of one serotype (Figure 1).
In our study, CIP resistance usually exists along with β-lactam resistance and ESBL production in the nine serotypes. Although there were several studies demonstrating concurrent resistance to CIP and cefotaxime in a particular serotype,25–27 due to the fact that these studies involved only one serotype, it is difficult to answer whether the concurrent resistance is mainly in a particular serotype or a strain or is a general case. This phenomenon proves that concurrent resistance to CIP and β-lactam is more a general phenomenon.
Finally, while analyzing the molecular mechanisms of resistance to CIP, the most interesting finding was that, rather than the commonly considered gyrA mutations, a type of PMQR structure qnrB is the most common indicator for CIP resistance because it was only found in all CIP-resistant isolates (Tables 4 and 5). The second most common indicator is aac(6 ¢)-Ib-cr. In contrast, gyrA mutations are not necessarily the cause of CIP resistance since these mutations were often found in reduced CIP-susceptible isolates and were not found in a number of CIP-resistant isolates. Especially, in reduced CIP-susceptible S. typhi, S83F and D87Y usually existed. Moreover, coexisting gyrA mutation and one or two PMQR genes were found in a number of reduced CIP-susceptible isolates, inconsistent with the previous statement that this coexistence is rare.25 To conclude, prevalence of qnrB and aac(6 ¢)-Ib-cr is significantly higher in CIP-resistant isolates, whereas gyr mutations in several CIP-resistant isolates (Table 4). Although it is usually believed that CIP resistance is mainly due to gyrA mutations28 and PMQR genes cause low-level resistance to CIP in enteric bacteria,29 our results demonstrate that it is not always the case in Salmonella. Given that it is difficult to obtain a large number of CIP-resistant isolates, we hope there will be more future studies to increase knowledge in this field. In addition, in highly CIP-resistant isolates (Kentucky and Indiana with MIC >16 μg/mL), presence of two gyrA mutations plus qnrB and/or aac(6′)-Ib-cr is common.
Conclusion
Our study shows that in Salmonella, there exists a high prevalence of reduced CIP susceptibility and a low prevalence of CIP resistance in limited serotypes. The CIP-resistant isolates generally show concurrent resistance to β-lactams. Our study also demonstrates that, rather than gyrA mutations, PMQR gene qnrB is the most common indicator for CIP resistance.
Acknowledgment
This study was supported by the Science and Technology Projects of Medicine and Health of Zhejiang (no. 2016KYB274 and 2018KY688).
Disclosure
The authors report no conflicts of interest in this work.
References
Majowicz SE, Musto J, Scallan E, et al; International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882–889. | ||
Centres for Disease Control and Prevention. Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2012 (Final Report). Atlanta, Georgia: U.S. Department of Health and Human Services, CDC; 2014. | ||
Centres for Disease Control and Prevention. An Atlas of Salmonella in the United States, 1968-2011: Laboratory-based Enteric Disease Surveillance. Atlanta, Georgia: US Department of Health and Human Services, CDC; 2013. | ||
Andersson MI, MacGowan AP. Development of the quinolones. J Antimicrob Chemother. 2003;51(suppl 1):1–11. | ||
Effa EE, Lassi ZS, Critchley JA, et al. Fluoroquinolones for treating typhoid and paratyphoid fever (enteric fever). Cochrane Database Syst Rev. 2011;(10):CD004530. | ||
Lo NW, Chu MT, Ling JM. Increasing quinolone resistance and multidrug resistant isolates among Salmonella enterica in Hong Kong. J Infect. 2012;65(6):528–540. | ||
Hassing RJ, Goessens WH, Mevius DJ, et al. Decreased ciprofloxacin susceptibility in Salmonella Typhi and Paratyphi infections in ill-returned travellers: the impact on clinical outcome and future treatment options. Eur J Clin Microbiol Infect Dis. 2013;32(10):1295–1301. | ||
Gong J, Wang C, Shi S, et al. Highly drug-resistant Salmonella enterica Serovar Indiana clinical isolates recovered from broilers and poultry workers with diarrhea in China. Antimicrob Agents Chemother. 2016;60(3):1943–1947. | ||
Westrell T, Monnet DL, Gossner C, Heuer O, Takkinen J. Drug-resistant Salmonella enterica serotype Kentucky in Europe. Lancet Infect Dis. 2014;14(4):270–271. | ||
Dutta P, Mitra U, Dutta S, De A, Chatterjee MK, Bhattacharya SK. Ceftriaxone therapy in ciprofloxacin treatment failure typhoid fever in children. Indian J Med Res. 2001;113:210–213. | ||
Fabrega A, Madurga S, Giralt E, Vila J. Mechanism of action of and resistance to quinolones. Microb Biotechnol. 2009;2(1):40–61. | ||
Giraud E, Baucheron S, Cloeckaert A. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 2006;8(7):1937–1944. | ||
Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25(5):358–373. | ||
Deak E, Hindler JA, Skov R, Sjolund-Karlsson M, Sokovic A, Humphries RM. Performance of Etest and disk diffusion for detection of ciprofloxacin and levofloxacin resistance in Salmonella enterica. J Clin Microbiol. 2015;53(1):298–301. | ||
CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard-Eleventh Edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. CLSI document M02-A12. | ||
CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. CLSI document M100-S27. | ||
Eaves DJ, Randall L, Gray DT, et al. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob Agents Chemother. 2004;48(10):4012–4015. | ||
Le TM, Baker S, Le TP, et al. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J Med Microbiol. 2009;58(pt 12):1585–1592. | ||
Centers for Disease Control and Prevention. Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Available from: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. Accessed May 4, 2016. | ||
Plant J, Glynn AA. Natural resistance to Salmonella infection, delayed hypersensitivity and Ir genes in different strains of mice. Nature. 1974;248(446):345–347. | ||
Cao TT, Deng GH, Fang LX, et al. Characterization of quinolone resistance in Salmonella enterica from farm animals in China. J Food Prot. 2017;80(10):1742–1748. | ||
Song Q, Yang Y, Lin W, Yi B, Xu G. Epidemiological characteristics and clinical treatment outcome of typhoid fever in Ningbo China, 2005-2014, PFGE results revealing great proportion of common transmission sources. Jpn J Infect Dis. 2017;70(5):513–516. | ||
Cui S, Li J, Sun Z, et al. Ciprofloxacin-resistant Salmonella enterica serotype Typhimurium, China. Emerg Infect Dis. 2008;14(3):493–495. | ||
Lu Y, Zhao H, Liu Y, et al. Characterization of quinolone resistance in Salmonella enterica serovar Indiana from chickens in China. Poult Sci. 2015;94(3):454–460. | ||
Bai L, Zhao J, Gan X, et al. Emergence and diversity of Salmonella enterica Serovar Indiana isolates with concurrent resistance to ciprofloxacin and cefotaxime from patients and food-producing animals in China. Antimicrob Agents Chemother. 2016;60(6):3365–3371. | ||
Wong MH, Yan M, Chan EW, Biao K, Chen S. Emergence of clinical Salmonella enterica serovar Typhimurium isolates with concurrent resistance to ciprofloxacin, ceftriaxone, and azithromycin. Antimicrob Agents Chemother. 2014;58(7):3752–3756. | ||
Wong MH, Zeng L, Liu JH, Chen S. Characterization of Salmonella food isolates with concurrent resistance to ceftriaxone and ciprofloxacin. Foodborne Pathog Dis. 2013;10(1):42–46. | ||
Chen S, Cui S, McDermott PF, et al. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob Agents Chemother. 2007;51(2):535–542. | ||
Liu JH, Deng YT, Zeng ZL, et al. Coprevalence of plasmid-mediated quinolone resistance determinants QepA, Qnr, and AAC(6’)-Ib-cr among 16S rRNA methylase RmtB-producing Escherichia coli isolates from pigs. Antimicrob Agents Chemother. 2008;52(8):2992–2993. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.