Back to Journals » OncoTargets and Therapy » Volume 12
Overexpression of miR-671-5p indicates a poor prognosis in colon cancer and accelerates proliferation, migration, and invasion of colon cancer cells
Received 13 June 2019
Accepted for publication 9 August 2019
Published 22 August 2019 Volume 2019:12 Pages 6865—6873
DOI https://doi.org/10.2147/OTT.S219421
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Wei Jin,1,* Jinsheng Shi,2,* Meiqin Liu1
1Department of Gastroenterology, Yidu Central Hospital of Weifang, Weifang, Shandong 262500, People’s Republic of China; 2Department of Pathology, Yidu Central Hospital of Weifang, Weifang, Shandong 262500, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Jin
Department of Gastroenterology, Yidu Central Hospital of Weifang, No.4138, South Linglongshan Road, Shandong 262500, People’s Republic of China
Tel +86 0 536 327 5615
Fax +86 0 536 327 9421
Email [email protected]
Purpose: Colon cancer is one of the common malignancies worldwide, and many genes, including microRNAs (miRNAs), have been demonstrated that associated with progression of various diseases, including cancers. The aim of this study is to investigate the potential role of miR-671-5p in colon cancer.
Patients and methods: Reverse transcription-quantitative polymerase chain reaction (qRT-PCR) was performed to detect the expression levels of miR-671-5p in 115 paired colon cancer tissues and adjacent normal tissues, as well as in colon cancer cells. Kaplan-Meier curve and Cox regression analyses were used to estimate the prognostic significance of miR-671-5p in colon cancer. CCK-8 assay, colony-formation assay, Transwell migration and invasion assays were used to evaluate the effects of miR-671-5p on cell proliferation, migration, and invasion in colon cancer.
Results: We found that miR-671-5p expression was increased in colon cancer tissues and cell lines. Overexpression of miR-671-5p was found associated with lymph node metastasis, TNM stage, and poor overall survival of patients with colon cancer. By exploiting miR-671-5p mimics and inhibitors, miR-671-5p overexpression significantly increased cell proliferation, migration, and invasion, while downregulation of miR-671-5p inhibited proliferation, migration, and invasion of colon cancer cells.
Conclusion: Taken together, miR-671-5p may act as an oncogene in colon cancer and promote proliferation, migration, and invasion of colon cancer cells by targeting TRIM67. And it may be a promising prognostic biomarker and therapeutic application for colon cancer treatment.
Keywords: miR-671-5p, colon cancer, prognosis, proliferation, migration, invasion
Introduction
Colon cancer is one of the most common gastrointestinal malignancies worldwide, with a high morbidity and mortality rate.1 Although the incidence and mortality rates of colon cancer have been declining in the United States, the incidence and mortality are still increasing in China, and colon cancer is a major public health problem.2,3 Metastasis is the leading cause of high mortality of colon cancer, especially the liver metastasis.4 At present, colon cancer patients at early stages have great opportunities to be cured by curative surgery treatment, however, the colon cancer patients with the advanced stages usually have a poor prognosis with the 5-year overall survival rate less than 60%.3,5,6 Thus, it is crucial to establish more effective biomarkers for colon cancer.
MicroRNAs (miRNAs) are a group of single-stranded, small, non-coding RNAs that post-transcriptional regulate mRNA. They usually negatively regulate target genes by direct interaction with the 3ʹ-untranslated region (3ʹ-UTR).7,8 Accumulating evidence has demonstrated numerous miRNAs are aberrantly expressed in various diseases, including cancers.9–11 The aberrant expression of miRNAs is involved in multiple biological processes, such as cell proliferation, differentiation, and migration, and associate with tumorigenesis and progression of cancers.12–14 A number of miRNAs have been reported to be upregulated or downregulated in colon cancer, such as miR-183,15 miR-137,16 and miR-302a.17 miR-671-5p has been found abnormal expressed in many cancers, such as pediatric chordomas,18 glioma,19 breast cancer,20 and advanced rectal cancer.21 However, the role of miR-671-5p in colon cancer is still elusive.
In the present study, the expression of miR-671-5p was detected in colon cancer tissues and cell lines. We also investigated whether miR-671-5p has clinical prognostic value in colon cancer. What’s more, we explored the effect of miR-671-5p on cell proliferation, migration, and invasion of colon cancer by upregulation or downregulation of miR-671-5p.
Materials and methods
Patient and tissue specimens
Between January 2010 and February 2013, a total of 115 paired colon cancer tissue specimens and adjacent normal tissue specimens (>5 cm from colon cancer tissues) were collected from 115 colon cancer patients during surgical resection in Yidu Central Hospital of Weifang. The tissue specimens were confirmed by two pathologists. These tissue specimens were immediately put into liquid nitrogen until use. None of the patients received radiotherapy or chemotherapy prior to surgical resection. All the patients signed written informed consent. The protocol was approved by the Ethics Committee of the Yidu Central Hospital of Weifang and conducted in accordance with the Declaration of Helsinki. Detailed clinicopathological characteristics and 5-year survival information of patients with colon cancer are collected and recorded.
Cell lines culture and transfection
The colon cancer cell lines SW480, SW620, LOVO, HCT116, and normal colon epithelial FHC cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). SW480 and SW620 were cultured in L15 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). HT29, HCT116, and FHC were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Nacalai Tesque, Japan) with 10% FBS. All cells were cultured at 37 °C in a humidified 5% CO2 atmosphere.
The miR-671-5p mimics (5ʹ-AGGAAGCCCUGGAGGGGCUGGAG-3ʹ), mimic negative control (mimic NC; 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-671-5p inhibitor (5ʹ-CUCCAGCCCCUCCAGGGCUUCCU-3ʹ), and inhibitor NC (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) were synthesized and purchased from the RiboBio (Guangzhou, China). Cells were seeded into 6-well plates with 50 nM of miR-671-5p mimic, mimic NC, miR-671-5p inhibitor, or inhibitor NC for transient transfection using the Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from all tissue specimens and cells by using TRIzol reagent extract (Thermo Fisher Scientific, Inc.) following the manufacturer’s protocol. A NanoDrop 2000 spectrophotometer was used to measure the extraction concentration. Then RNA was transcribed into cDNA using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qRT-PCR analysis was carried out to detect the expression levels of miR-671-5p using SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific, Inc.). The relative expression of miR-671-5p was calculated using 2−ΔΔCt method with U6 used as an internal control.
CCK-8 assay
The proliferation of colon cancer cells was measured using the Cell Counting Kit-8 (CCK-8, Beyotime Institute). After transfection, colon cancer cells (3×103 cells/well) were seeded in 96-well plates and cell proliferation capacities were measured at 0, 24, 48, and 72 h. 10 μl CCK-8 reagent was added into culture medium per well at each time point and the optical density of solution was measured after 1 h using a microplate reader at 450 nm.
Colony-formation ability assay
The colony-forming ability of colon cancer cells was evaluated by colony-formation ability assays. For the colony formation assay, transfected cells (400 cells/well) were seeded into 6-well culture dishes and cultured in complete medium at 37 °C in 5% CO2 for 14 days. The cells were washed with PBS and stained with crystal violet for 15 min. Visible colonies were counted and imaged.
Transwell migration and invasion assays
The migration and invasion capacities of colon cancer cells were measured using Transwell plates (Corning Life Sciences, NY, USA) with 8 μm pore size membranes. For cell migration assay, the membranes are without Matrigel, but for cell invasion assay, membranes are pre-coated with Matrigel (Corning, USA). 3×104 transfected cells in FBS-free culture medium were seeded in the upper chambers while the culture medium containing 10% FBS was placed in the lower chambers as the attractant. After 24 h of incubation at 37 °C with 5% CO2, the cells on the upper chamber membranes were removed, whereas migrated or invasive cells on the lower chamber membranes were fixed and stained. The number of the cells was counted with a light microscope (Olympus Corporation, Tokyo, Japan).
Bioinformatics analysis and dual-luciferase reporter assay
Targetscan (http://www.targetscan.org/) and miRDB (http://www.mirdb.org/) were used to predict the target sites of miR-671-5p through bioinformatics analysis. The TRIM67 3ʹ-UTR was identified at miR-671-5p binding sites, and wild-type and mutant TRIM67 3ʹ-UTR luciferase reporter gene plasmids were inserted into the pGL3 luciferase reporter vector (Promega, USA) to generate TRIM67 3ʹ-UTR wide-type (WT) or TRIM67 3ʹ-UTR mutant (MUT). Then the reporter vectors (TRIM67 3ʹ-UTR WT or TRIM67 3ʹ-UTR MUT) and miR-671-5p mimic, mimic NC, miR-671-5p inhibitor, or inhibitor NC were cotransfected into LOVO cells using Lipofectamine 3000 reagent (Invitrogen) and cultivated for 48 h for luciferase reporter assay. The relative luciferase activities were normalized to Renilla luciferase activities.
Statistical analysis
All statistical analyses were analyzed using the SPSS 20.0 statistical software (SPSS Inc., Chicago, IL, USA) and GraphPad 5.0 software (GraphPad Software, Inc., USA). Data are presented as the mean ± SD. Differences were compared by using the χ2 test, paired Student’s t-test, or one-way ANOVA. The Kaplan-Meier curve and Cox regression analyses were used to assess the prognostic value of miR-671-5p in colon cancer. All the experiments in this study were performed at least three times. It is considered to be statistically significant when P<0.05.
Results
Expression of miR-671-5p in colon cancer tissues and cells
First, we detected the expression levels of miR-671-5p in a total of 115 pairs of colon cancer tissue specimens and adjacent normal tissue specimens by qRT-PCR. As shown in Figure 1A, miR-671-5p levels in colon cancer tissues were significantly higher compared with the expression in adjacent normal tissues (P<0.001). Next, we examined the expression levels of miR-671-5p in colon cancer cell lines. Compared to the expression levels in the normal colon epithelial cell line FHC, miR-671-5p has higher expression levels in colon cancer cell lines, including SW480, SW620, LOVO, and HCT116 (P<0.001, Figure 1B).
miR-671-5p has relatively higher expression levels in SW620 and LOVO cell lines compared with the other two colon cancer cell lines (SW480 and HCT116) as indicated in Figure 1B. SW620 and LOVO cell lines were therefore selected for subsequent study.
Association between miR-671-5p expression and clinical characteristics of colon cancer patients
We further analyzed the association between miR-671-5p expression and clinical characteristics of patients with colon cancer. In order to facilitate the analysis, colon cancer patients were stratified by the relative mean expression level (3.838) of miR-671-5p in colon cancer tissue specimens. The results in Table 1 showed that miR-671-5p expression is significantly associated with lymph node metastasis (P=0.013) and TNM stage (P=0.005). However, no significant difference is observed relating to other clinical parameters, such as gender, age, tumor size, and differentiation (P>0.05, Table 1).
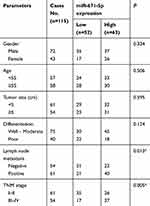 |
Table 1 Relation between miR-671-5p expression levels and clinical characteristics in patients with colon cancer |
The miR-671-5p expression is associated with the overall survival time of colon cancer patients
Considering that miR-671-5p expression is significantly associated with lymph node metastasis and TNM stage, we used Kaplan-Meier curve and Cox regression analyses to assess whether the miR-671-5p expression was associated with prognosis of colon cancer patients. The Kaplan-Meier survival analysis results showed that colon cancer patients with high levels of miR-671-5p had significantly shorter overall survival time than those with low levels of miR-671-5p (log-rank P=0.001, Figure 2). The multivariate Cox analysis results suggested that high levels of miR-671-5p (HR =2.572, 95% CI: 1.247–5.304, P=0.011, Table 2) and TNM stage (HR =2.317, 95% CI: 1.184–4.534, P=0.014, Table 2) were independent prognostic risk factors for colon cancer patients.
 |
Table 2 Multivariate Cox analysis for the association between clinical characteristics and overall survival of colon cancer patients |
 |
Figure 2 Kaplan-Meier curve showed that overexpression of miR-671-5p is associated with shorter overall survival time of colon cancer patients. |
miR-671-5p promotes colon cancer cell proliferation, migration, invasion in vitro
To further explore the role of miR-671-5p in colon cancer, SW620 and LOVO cells were transfected with miR-671-5p mimic, miR-671-5p inhibitor, or respective negative controls. After transfection, qRT-PCR was used to verify the efficiency of transfection. The results showed that following transfection of miR-671-5p mimic in SW620 and LOVO cancer cells, the expression of miR-671-5p was significantly increased, while in those cells transfected with miR-671-5p inhibitor, the expression of miR-671-5p was dramatically decreased (P<0.01. Figure 3A). The CCK-8 assay was performed to evaluate the proliferation capacity of miR-671-5p in SW620 and LOVO cancer cells. Results revealed that overexpression or downregulation of miR-671-5p promoted or inhibited colon cancer cell proliferation, respectively, compared with that in untreated cells (P<0.05, Figure 3B). Colony formation assays showed that overexpression of miR-671-5p promoted colony formation, while downregulation of miR-671-5p inhibited colony formation, compared with that in untreated cells (P<0.05, Figure 3C). Subsequently, Transwell migration and invasion assays showed that overexpression or knockdown of miR-671-5p significantly promoted or inhibited the migration or invasion abilities of colon cancer cells, respectively (P<0.01, Figure 4A and B). The above results indicate that miR-671-5p could promote colon cancer progression in vitro.
TRIM67 was a direct target gene for miR-671-5p
We searched the potential targets of miR-671-5p using bioinformatics analysis. Among these candidate target genes, the TRIM67 3ʹ-UTR was identified containing a putative miR-671-5p binding sites (Figure 5A) and TRIM67 was subsequently selected for further examination, as it functioned as a tumor suppressor in colorectal cancer to suppress colorectal cancer initiation and progression.22 Then we used luciferase reporter assay to confirm this prediction, as presented in Figure 5B, the luciferase activity of the reporter containing WT TRIM67 3ʹ-UTR was significantly suppressed in the cells overexpression miR-671-5p (P<0.05), while there was no change in luciferase activity of the reporter containing the mutant binding site, compared with untreated cells. These findings indicated that TRIM67 may be a direct target of miR-671-5p in colon cancer.
Discussion
In this study, we found that high miR-671-5p was associated with some clinical characteristics of colon cancer patients, as well as poor prognosis of colon cancer patients. This study further revealed that high expression of miR-671-5p may promote the progression of colon cancer by targeting TRIM67.
Despite advances in treatment for colon cancer and liver metastasis, and the survival rate for patients has largely improved in the United States, however, the prognosis of patients with colon cancer is still unsatisfactory in China.2,23 In recent decades, increasing evidence has shown that miRNAs are aberrantly expressed and function as diagnostic and/or prognostic biomarkers in various cancers, as well as play crucial roles in tumor progression.9,24,25 For instance, miR-17 is found upregulated in breast cancer and can be used as a prognostic biomarker and promote cell proliferation in breast cancer.26 In a systematic review study, low miR-375 expression was associated with significantly poorer outcomes and it could act as a valuable prognostic marker in various cancers.27 These studies underscore the importance of finding more effective biomarkers associated with the progression of cancers.
Previous studies have indicated that some miRNAs are associated with the tumorigenesis, development of colon cancer by affecting cancer cell behaviors, such as miR-21, miR-138, miR-155, and miR-873-5p.28–30 miR-21 and miR-138 are found aberrantly expressed in colon cancer tissues and high miR-21 expression is associated with the degree of malignancy of patients while low miR-138 expression is positively associated with survival, both of which may be involved in the regulation of colon cancer cell proliferation.28 The expression of miR-155 is upregulated in colonic cancer tissues and it may act as proto-oncogenes related to carcinogenesis and development of colon cancer.29 miR-873-5p is found downregulated in colon cancer and functions as a tumor suppressor to inhibit the progression of colon cancer via repression of TUSC3/AKT signaling.30
Dysregulated miR-671-5p is reported in several types of human cancers.18–20 In the present study, we investigated the expression levels of miR-671-5p in colon cancer tissues and cell lines by using qRT-PCR. The results demonstrated that miR-671-5p was upregulated in both colon cancer tissues and cell lines. Overexpression of miR-671-5p was strongly associated with lymph node metastasis and TNM stage of patients with colon cancer. These results implied that miR-671-5p may be involved in the tumorigenesis and development of colon cancer. Considering miR-671-5p expression was significantly associated with lymph node metastasis and TNM stage, we used Kaplan-Meier curve and Cox regression analyses to investigate whether miR-671-5p expression has prognostic significance in colon cancer. The results indicated that high miR-671-5p was associated with a poor survival rate of colon cancer patients and it might be an independent prognostic factor for colon cancer patients in the present study.
To further explore whether miR-671-5p act as an oncogenic role in colon cancer, cell proliferation, migration, and invasion abilities were investigated in colon cancer cells by upregulating or downregulating miR-671-5p. It was identified that miR-671-5p promoted cell proliferation, migration, and invasion. Hence, these results confirmed the oncogenic effects of miR-671-5p on regulating colon cancer progression in the current study. These results are consistent with the results of miR-671-5p in several other cancers. For instance, miR-671-5p was upregulated in prostate cancer and promoted prostate cancer cell proliferation by inhibiting SOX6.31 Malgulwar et al suggested that upregulation of miR-671-5p and miR-193-5p expressions mediated epigenetic mode of SMARCB1/INI1 loss and downregulated TGF-β pathway in childhood chordomas.18 In glioblastoma multiforme (GBM), miR-671-5p was found overexpressed in both biopsies and cell lines, and overexpression of miR-671-5p significantly increased migration of GBM cells through the axis miR-671-5p/CDR1-AS/CDR1/VSNL1.19 These above studies have also validated few genes as direct targets of miR-671-5p. In the present study, bioinformatics analysis predicted TRIM67 as a potential target of miR-671-5p. The luciferase reporter assays further demonstrated that miR-671-5p directly targeted the TRIM67 3ʹUTR. TRIM67 belongs to the class I-2 of TRIM protein family that is a highly conserved super-family of E3 ubiquitin ligases.32 In recent years, TRIM67 has demonstrated involved in a number of critical processes and cancer.33–35 For instance, TRIM67 promotes the processing of NF-κB2 into its active form and cell apoptosis in GA-13315-treated lung cancer cells.36 In colorectal cancer, TRIM67 functions as a pivotal tumor suppressor to suppress colorectal cancer initiation and progression.22 Therefore, we speculate that miR-671-5p may promote cell proliferation, migration, and invasion by targeting TRIM67. In the current study, the results observed that miR-671-5p may be involved in the development and progression of colon cancer by targeting TRIM67. However, there are some limitations in our present study. The sample size was small, which may affect our conclusions. Further studies are needed to explore the potential role and detailed mechanism of miR-671-5p in colon cancer.
Taken together, miR-671-5p is upregulated in colon cancer tissues and cell lines, as well as associated with the survival rate of colon cancer patients. miR-671-5p may serve a role as an oncogene in colon cancer by promoting cell proliferation, migration, and invasion in vitro by targeting TRIM67. Therefore, the present study suggested the theoretical basis of miR-671-5p for the prognosis and treatment of colon cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi:10.1158/1055-9965.EPI-15-0578
2. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi:10.3322/caac.21395
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
4. Worni M, Shah KN, Clary BM. Colorectal cancer with potentially resectable hepatic metastases: optimizing treatment. Curr Oncol Rep. 2014;16:407. doi:10.1007/s11912-014-0407-z
5. Freeman HJ. Early stage colon cancer. World J Gastroenterol. 2013;19:8468–8473. doi:10.3748/wjg.v19.i46.8468
6. Wang Y, He S, Zhu X, Qiao W, Zhang J. High copy number of mitochondrial DNA predicts poor prognosis in patients with advanced stage colon cancer. Int J Biol Markers. 2016;31:e382–e388. doi:10.5301/jbm.5000211
7. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi:10.1146/annurev.cellbio.23.090506.123406
8. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. doi:10.1055/s-0034-1397344
9. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi:10.1038/nrd.2016.246
10. Wahid F, Khan T, Kim YY. MicroRNA and diseases: therapeutic potential as new generation of drugs. Biochimie. 2014;104:12–26. doi:10.1016/j.biochi.2014.05.004
11. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013
12. Yoshikawa T, Wu J, Otsuka M, et al. Repression of MicroRNA function mediates inflammation-associated colon tumorigenesis. Gastroenterology. 2017;152:631–643. doi:10.1053/j.gastro.2016.10.043
13. Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15:1444–1455. doi:10.4161/15384047.2014.955442
14. Liang T, Hu XY, Li YH, Tian B-Q, Li Z-W, Fu Q. MicroRNA-21 regulates the proliferation, differentiation, and apoptosis of human renal cell carcinoma cells by the mTOR-STAT3 signaling pathway. Oncol Res. 2016;24:371–380. doi:10.3727/096504016X14685034103356
15. Bi DP, Yin CH, Zhang XY, Yang -N-N, Xu J-Y. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep. 2016;35:2873–2879. doi:10.3892/or.2016.4631
16. Guo Y, Pang Y, Gao X, et al. MicroRNA-137 chemosensitizes colon cancer cells to the chemotherapeutic drug oxaliplatin (OXA) by targeting YBX1. Cancer Biomark. 2017;18:1–9. doi:10.3233/CBM-160650
17. Sun S, Zhang G, Wu Z, et al. MicroRNA-302a functions as a putative tumor suppressor in colon cancer by targeting Akt. PLoS One. 2014;9:e115980. doi:10.1371/journal.pone.0115980
18. Malgulwar PB, Pathak P, Singh M, et al. Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain Tumor Pathol. 2017;34:155–159. doi:10.1007/s10014-017-0295-7
19. Barbagallo D, Condorelli A, Ragusa M, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7:4746–4759. doi:10.18632/oncotarget.6621
20. Tan X, Fu Y, Chen L, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7:293–307. doi:10.18632/oncotarget.6344
21. Della Vittoria Scarpati G, Falcetta F, Carlomagno C, et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113–1119. doi:10.1016/j.ijrobp.2011.09.030
22. Wang S, Zhang Y, Huang J. et al. TRIM67 activates p53 to suppress colorectal cancer initiation and progression. Cancer Res;2019.
23. Zhang Y, Shi J, Huang H, et al. [Burden of colorectal cancer in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2015;36:709–714.
24. Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93. doi:10.1016/j.addr.2014.09.001
25. Luu HN, Lin HY, Sorensen KD, et al. miRNAs associated with prostate cancer risk and progression. BMC Urol. 2017;17:18. doi:10.1186/s12894-017-0206-6
26. Yang F, Li Y, Xu L, et al. miR-17 as a diagnostic biomarker regulates cell proliferation in breast cancer. Onco Targets Ther. 2017;10:543–550. doi:10.2147/OTT.S127723
27. Dan B, Luo J, Li K, Chen S. Prognostic value of miR-375 for survival outcomes in various cancers: a systematic review and meta-analysis. Oncol Res Treat. 2018;41:47–50. doi:10.1159/000481708
28. You C, Jin L, Xu Q, Shen B, Jiao X, Huang X. Expression of miR-21 and miR-138 in colon cancer and its effect on cell proliferation and prognosis. Oncol Lett. 2019;17:2271–2277. doi:10.3892/ol.2018.9864
29. Cao H, Huang S, Liu A, Chen Z. Up-regulated expression of miR-155 in human colonic cancer. J Cancer Res Ther. 2018;14:604–607. doi:10.4103/0973-1482.175432
30. Zhu Y, Zhang X, Qi M, Zhang Y, Ding F. miR-873-5p inhibits the progression of colon cancer via repression of TUSC3/AKT signaling. J Gastroenterol Hepatol. 2019. doi:10.1111/jgh.14697
31. Yu Y, Wang Z, Sun D, et al. miR-671 promotes prostate cancer cell proliferation by targeting tumor suppressor SOX6. Eur J Pharmacol. 2018;823:65–71. doi:10.1016/j.ejphar.2018.01.016
32. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi:10.1016/j.tibs.2017.01.002
33. Zhan W, Han T, Zhang C, et al. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One. 2015;10:e0142596. doi:10.1371/journal.pone.0142596
34. Boyer NP, Monkiewicz C, Menon S, Moy SS, Gupton SL. Mammalian TRIM67 functions in brain development and behavior. eNeuro. 2018;5. doi:10.1523/ENEURO.0186-18.2018
35. Do LD, Gupton SL, Tanji K, et al. TRIM9 and TRIM67 are new targets in paraneoplastic cerebellar degeneration. Cerebellum. 2019;18:245–254. doi:10.1007/s12311-018-0987-5
36. Liu R, Chen Y, Shou T, Hu J, Chen J, Qing C. TRIM67 promotes NFkappaB pathway and cell apoptosis in GA13315treated lung cancer cells. Mol Med Rep. 2019;20:2936–2944. doi:10.3892/mmr.2019.10509
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




