Back to Journals » OncoTargets and Therapy » Volume 13
Overexpression of LncRNA SNHG1 Were Suitable for Oncolytic Adenoviruse H101 Therapy in Oral Squamous-Cell Carcinoma
Authors Wang X, Yang S, Lv X, Wang L, Li C
Received 16 October 2020
Accepted for publication 20 November 2020
Published 21 December 2020 Volume 2020:13 Pages 13033—13039
DOI https://doi.org/10.2147/OTT.S285536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Xin Wang, 1,* Song Yang, 1,* Xuechao Lv, 2 Lina Wang, 3 Chunmei Li 4
1Department of Plastic Maxillofacial Surgery, Heilongjiang Provincial Hospital, Harbin 150036, Heilongjiang, People’s Republic of China; 2Department of Pediatric Dentistry, School of Stomatology, Harbin Medical University, Harbin 150001, Heilongjiang, People’s Republic of China; 3Department of Oral and Maxillofacial Surgery, Second Affiliated Hospital of Harbin Medical University, Harbin 150086, Heilongjiang, People’s Republic of China; 4Department of Outpatient Stomatology, Heilongjiang Provincial Hospital, Harbin 150036, Heilongjiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunmei Li
Department of Outpatient Stomatology, Heilongjiang Provincial Hospital, Harbin 150036, Heilongjiang, People’s Republic of China
Tel +86 451-8713-1023
Email [email protected]
Background: As the most prevalent type of head and neck cancer, oral squamous-cell carcinoma (OSCC) accounts for nearly 90% of all oral cancer cases. Despite great progress having been made in the diagnosis and treatment of OSCC recently, the survival rate of OSCC patients has not risen remarkably. Chemotherapy is commonly used for OSCC treatment; however, the emergence of chemoresistance limits its long-term curative effect. Therefore, identifying effective biomarkers and molecular mechanisms is essential to the development of therapeutic strategies for OSCC.
Methods: qRT-PCR assays were performed to detect SNHG1 expression in OSCC tissue and cells, and CCK8 assays and animal experiments used to examine cell proliferation. In addition, CCK8 assays were used to detect IC 50 values of cisplatin, 5Fu, Dox, and oncolytic adenovirus H101.
Results: We found that SNHG1 was overexpressed in OSCC tissue and cells and was associated with OSCC progression. In addition, knockdown of SNHG1 suppressed cell proliferation in vitro and in vivo. Importantly, we found that oncolytic adenovirus H101 showed better antitumor effects in OSCC with high SNHG1 expression, and chemotherapy showed worse anti-tumor effects in OSCC with high SNHG1 expression.
Conclusion: SNHG1 can act as a diagnostic biomarker for OSCC, and may be a biomarker for treatment options.
Keywords: SNHG1, H101, OSCC
Introduction
As the most prevalent type of head and neck cancer, oral squamous-cell carcinoma (OSCC), accounts for nearly 90% of all oral cancer cases.1–3 Despite great progress having been made in the diagnosis and treatment of OSCC recently, the survival rate of OSCC patients has not risen remarkably.4–6 Therefore, identifying effective biomarkers and molecular mechanisms is essential to the development of therapeutic strategies for OSCC.
Lacking protein-coding capacity, lncRNAs are a class of regulatory transcripts >200 nucleotides. Early research revealed lncRNAs played a significant role in tumorigenesis via multiple mechanisms, such as with microRNAs or proteins. For example, DANCR lncRNA promotes tumor progression and cancer-stemness features in osteosarcoma by upregulating Axl via miR33a-5p inhibition.7 SNHG3 lncRNA induces epithelial–mesenchymal transition and sorafenib resistance by modulating the miR128–CD151 pathway in hepatocellular carcinoma.8 CRNDE lncRNA promotes colorectal cancer–cell proliferation and chemoresistance via miR181a-5p–mediated regulation of Wnt–β-catenin signaling.9 Therefore, study of lncRNAs may be of remarkable value in understanding the occurrence and development of tumors.
In the exploration for therapeutic strategies for tumor, oncolytic viruses have recently displayed potential and effective antitumor effects.10,11 For example, there is a novel approach to glioma therapy using an oncolytic adenovirus with two specific promoters.12 An oncolytic adenovirus enhances antiangiogenic and antitumoral effects of a replication-deficient adenovirus encoding endostatin by rescuing its selective replication in nasopharyngeal carcinoma cells.13 Oncolytic adenovirus expressing IL18 induces significant antitumor effects against melanoma in mice through inhibition of angiogenesis.14
Recently found, SNHG1 lncRNA is located at chromosome 11q12.3. SNHG1 has been demonstrated to acts as an oncogene incolon cancer,15–20 hepatocellular carcinoma, lung cancer, osteosarcoma, neuroblastoma, and cholangiocarcinoma. However, until now, no report has revealed the function of SNHG1 in the progression and development of OSCC.
In this study, we found that SNHG1 was remarkably increased in OSCC tissue and associated with tumor progression. Moreover, we found that oncolytic adenovirus H101 was more suitable for therapy of OSCC with high SNHG1 expression and chemotherapy more suitable for therapy of OSCC with low SNHG1 expression. As such, SNHG1 can act as a diagnostic biomarker for OSCC and might be a biomarker for treatment options.
Methods
Clinical Samples
A total of 152 FFPE block samples consisting of 76 paraffin blocks of OSCC tissue and 76 paraffin blocks of nontumoral tissue collected from the Department of Outpatient Stomatology, Heilongjiang Hospital were used. None of the patients had received chemotherapy or radiotherapy before surgery. This study was approved by the Ethics Committee of Heilongjiang Hospital. All patients provided written informed consent in accordance with the ethical guidelines of Heilongjiang Hospital, and the research conformed with good clinical practice and the principles laid out in the Declaration of Helsinki.
Cell Culture
The OSCC cell lines SCC9, SCC25, HN4, and Cal27 and the hNOK cell line were purchased from the Shanghai Model Cell Bank (http://www.cellbank.org.cn/mulu.asp). These cells were cultured inDMEM; (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% FBS,and 100 µg/mL penicillin–streptomycin (BioLight, Shanghai, China) at 37°C with 5% CO2. To knock down SNHG1, SCC25 or HN4 cells (2×105) were seeded in six-well dishes at 24 hours and then transfected with SNHG1, sh-NC, or sh-SNHG1 per dish using Lipofectamine 3000 according to the manufacturer’s instructions. Sequences for sh-SNHG1 (Hanheng Biotechnology) were 5ʹ- CCGGCCTTGGGTCTGGAAACTGTTACTCGAGTAACAGTTTCCAGACCCAAGGTTTTTTG-3ʹ, and those for sh-NC (Hanheng Biotechnology) 5ʹ-CCGGATCTCGCTTGGGCGAGAGTAAGTCGACTTACTCTCGCCCAAGCGAGATTTTTTTG-3ʹ.
RNA Extraction and qRT-PCR
Total RNA was isolated using an RNeasy minikit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions. To examine SNHG1 expression, PrimeScript RT master mix (Takara Bio, Kusatsu, Japan) and 2× PCR Master Mix (Thermo Fisher Scientific) were used for reverse transcription (RT) and qPCR, respectively. The relative expression of SNHG1 was calculated and normalized using the 2–ΔΔCt method. Primer sequences used were: SNHG1 forward 5ʹ-AGGCTGAAGTTACAGGTC-3ʹ, reverse 5ʹ-TTGGCTCCCAGTGTCTTA-3ʹ, GAPDH forward 5ʹ-GGAGCGAGATCCCTCCAAAAT-3, and reverse 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ.
Cell Proliferation
Equal numbers of transfected OSCC cells (5,000 cells/well) were seeded into a 96-well plate and cell proliferation detected using CCK8 (Biyuntian Biotechnology, Shanghai, China) every 24 hours in accordance with the manufacturer’s protocols. A microplate reader was used to examine optical density at 450 nm.
IC50 Assays
Equal numbers of transfected OSCC cells (5,000 cells/well) were seeded in a 96-well plate and incubated with increasing concentrations of DDP (0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, 12.8, and 25.6 µg/mL), Dox (0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, 12.8, and 25.6 µg/mL), 5Fu (0.025, 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4, 12.8, and 25.6 µg/mL), and H101 (0.0625, 0.125, 0.25, 0.5, 1, 5, 10, 20, 40, and 80 MOI) for another 48 hours. Subsequently, IC50 was detected usingCCK8 (Biyuntian Biotechnology, Shanghai, China) in accordance with the manufacturer’s protocols. A microplate reader was used to examine optical density at 490 nm.
Animal Experiments
To detect OSCC-cell proliferation in vivo, ten BALB/c nude male mice (4–6 weeks old) were purchased from the Shanghai Laboratory Animal Center (Shanghai, China) and divided into two groups (five in each group). NH4-sh-NC (5×106) or NH4-sh-SNHG1 cells (5×106) were injected subcutaneously into the flanks of nude mice and tumor volumes measured every 7 days according to the formula: 0.5 × length × width2. All animal experiments were performed according to the Guide for the Care and Use of Laboratory Animals and the ethical guidelines of Heilongjiang Hospital. This work was approved by the Ethics Committee of Heilongjiang Hospital.
Statistical Analysis
GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. Differences in SNHG1 expression in OSCC tissue among patients were analyzed using the χ2 test for the clinical parameters as age, sex, smoking, TNM stage, and lymph-node metastasis. Log-rank (Mantel–Cox) tests, one‐way ANOVA, ROC curves, and Student’s t-tests were used for statistical analysis, and quantitative data are presented as means ± SD. Results were considered statistically significant at P<0.05.
Results
SNHG1 Overexpressed in OSCC Tissue
Using qRT-PCR, we examined SNHG1 expression in both OSCC and normal tissue. This showed that SNHG1 expression was significantly increased in OSCC tissue compared to normal tissue (Figure 1), which was consistent with previous research, suggesting that aberrant SNHG1 expression might be associated with OSCC progression and development.
 |
Figure 1 SNHG1 was overexpressed in OSCC tissue (n=76.) *p<0.05). All experiments were repeated three times. |
Clinical Significance of SNHG1 in Indicating OSCC Progression
Relationships between SNHG1 expression and clinicopathological characteristics of OSCC were studied. As summarized in Table 1, high SNHG1 expression was related to lymph-node metastasis, TNM stage, and tumor size (P<0.05). However, there were no significant relationships for sex, age, or smoking (P>0.05). Moreover, patients with high SNHG1 expression had poor survival when compared to patients with low SNHG1 expression (Figure 2A), and SNHG1 expression showed potential clinical significance as a tumor biomarker on the ROC curve (Figure 2B).
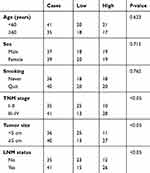 |
Table 1 Correlations Between SNHG1 Expression and Clinicopathological Characteristics in 76 Patients with OSCC |
Knockdown of SNHG1 Suppressed Cell Proliferation in OSCC
To investigate the role of SNHG1 in the progression and development of OSCC, we examined SNHG1 expression in OSCC cells. As shown in Figure 3A, SNHG1 expression was remarkably upregulated in OSCC cells compared to hNOK cells. Moreover, qRT-PCR assays revealed that SNHG1 expression was decreased in OSCC cells transfected with SNHG1 shRNA (Figure 3B), suggesting that we had successfully achieved stable knockdown of SNHG1. CCK8 assays showed that knockdown of SNHG1 significantly inhibited proliferation of OSCC cells (Figure 3C and D). In addition, we found that SNHG1 knockdown markedly suppressed proliferation of OSCC cells in vivo (Figure 3E), and mice having received SNHG1 knockdown had great survival compared to the control group (Figure 3F). Therefore, knockdown of SNHG1 suppressed cell proliferation in OSCC.
High SNHG1 Expression in OSCC Suitable for H101 Therapy
Oncolytic viruses have recently displayed potentially effective antitumor effects. qRT-PCR assays showed that SNHG1 expression was increased in OSCC cells transfected with SNHG1 and decreased in OSCC cells transfected with sh-SNHG1 (Figure 4A). In addition, CCK8 assays revealed that IC50 values for cisplatin (Figure 4B), Dox (Figure 4C), and 5Fu (Figure 4D) increased with higher expression of SNHG1 in OSCC cells, while H101 IC50 decreased with higher expression of SNHG1 in OSCC cells (Figure 4E). Therefore, H101 exhibited a stronger antitumor effect in OSCC cells with high SNHG1 expression, and chemotherapy exhibited a stronger antitumor effect in OSCC cells with low SNHG1 expression.
Discussion
Emerging lines of evidence indicate that lncRNAs function as oncogenes or tumor-suppressor genes to regulate carcinogenesis, and that they can be used as diagnostic or prognostic markers.21 For example, a new tumor suppressor — ADAMTS9-AS2 lncRNA — is regulated by DNMT1 and inhibits migration of glioma cells.22 -FEZF1-AS1 lncRNA promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling.23 LncRNA-135528 inhibits tumor progression by upregulating CXCL10 through the JAK–STAT pathway.24 In this work, the SNHG1 expression was remarkably upregulated in OSCC cells and tissue, and its overexpression was associated with advanced stage and unfavorable prognosis of OSCC patients. Moreover, SNHG1 knockdown suppressed cell proliferation in vitro and in vivo. Similarly, SNHG1 lncRNA indicates a poor prognosis and promotes colon cancer tumorigenesis.15 Likewise, SNHG1 lncRNA indicates a poor prognosis and promotes hepatocellular carcinoma tumorigenesis.16 Upregulated lncRNA SNHG1 contributes to progression of non–small cell lung cancer through inhibition of miR101-3p and activation of the Wnt–β-catenin signaling pathway.17 As such, our data indicated the oncogenic role of SNHG1 in the development of OSCC.
How to choose a treatment suitable for OSCC is still unclear. Increasingly, studies have indicated that oncolytic viruses have potential and effective antitumor effects.10,11 Chemotherapy is commonly used for OSCC treatment; however, the emergence of chemoresistance limits its long-term curative effect. lncRNA plays a significant role in OSCC chemoresistance.25,26 Therefore, we ran IC50 assays to examine the sensitivity of oncolytic adenoviruses and chemotherapy drugs to OSCC so as to determine the role of SNHG1 in tumor treatment. We found that oncolytic adenovirus H101 exhibited higher sensitivity in OSCC with high SNHG1 expression and chemotherapy exhibited higher sensitivity in OSCC with low SNHG1 expression. The occurrence and development of tumors involve multiple genes, so in the process of treatment, the selection of highly targeted treatment strategies can achieve more effective therapy. We have identified SNHG1 as a biomarker for OSCC therapy, and our research may help to establish a therapeutic strategies by detecting of SNHG1 expression.
In summary, this research identified the expression and functional role of SNHG1 in OSCC. Our results revealed that SNHG1 was remarkably increased in OSCC tissue and cells. SNHG1 was associated with advanced stage and unfavorable prognosis of OSCC patients, and thus acted as an oncogenic in OSCC. Moreover, knockdown of SNHG1 suppressed cell proliferation in vitro and in vivo. Importantly, we found that oncolytic adenovirus H101 was more suitable for OSCC therapy with high SNHG1 expression and chemotherapy more suitable OSCC therapy with low SNHG1 expression. In future, we will further study the relationship between the expression of SNHG1 in serum and chemotherapy and oncolytic virus therapy to ensure that the OSCC-treatment process can provide faster and more effective treatment options. SNHG1 can act as a diagnostic biomarker for OSCC, and may be expected to be a biomarker for treatment options.
Acknowledgments
This study was supported by the Heilongjiang Natural Science Foundation (Face project; H2017042).
Disclosure
The authors declare no conflicts of interest.
References
1. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–316. doi:10.1016/j.oraloncology.2008.06.002
2. Kademani D. Oral cancer. Mayo Clin Proc. 2007;82(7):878–887. doi:10.4065/82.7.878
3. Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52(4):195–215.
4. Vaishampayan UN, Podgorski I, Heilbrun LK, et al. Biomarkers and bone imaging dynamics associated with clinical outcomes of oral cabozantinib therapy in metastatic castrate-resistant prostate cancer. Clin Cancer Res. 2019;25(2):652–662. doi:10.1158/1078-0432.CCR-18-1473
5. Hsieh MY, Chen G, Chang DC, Chien SY, Chen MK. The impact of metronomic adjuvant chemotherapy in patients with advanced oral cancer. Ann Surg Oncol. 2018;25(7):2091–2097.
6. Hoffman HT, Porter K, Karnell LH, et al. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116(9Pt 2 Suppl 111):1–13. doi:10.1097/01.mlg.0000236095.97947.26
7. Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi:10.1016/j.canlet.2017.06.009
8. Zhang PF, Wang F, Wu J, et al. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. 2019;234(3):2788–2794. doi:10.1002/jcp.27095
9. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1
10. Nemunaitis J, Edelman J. Selectively replicating viral vectors. Cancer Gene Ther. 2002;9(12):987–1000.
11. Geoerger B, Vassal G, Opolon P, et al. Oncolytic activity of p53-expressing conditionally replicative adenovirus AdDelta24-p53 against human malignant glioma. Cancer Res. 2004;64(16):5753–5759. doi:10.1158/0008-5472.CAN-04-0499
12. Liu F, Xu K, Yang H, et al. A novel approach to glioma therapy using an oncolytic adenovirus with two specific promoters. Oncol Lett. 2018;15(3):3362–3368.
13. Liu RY, Zhou L, Zhang YL, et al. An oncolytic adenovirus enhances antiangiogenic and antitumoral effects of a replication-deficient adenovirus encoding endostatin by rescuing its selective replication in nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;442(3–4):171–176. doi:10.1016/j.bbrc.2013.11.047
14. Zheng JN, Pei DS, Mao LJ, et al. Oncolytic adenovirus expressing interleukin-18 induces significant antitumor effects against melanoma in mice through inhibition of angiogenesis. Cancer Gene Ther. 2010;17(1):28–36. doi:10.1038/cgt.2009.38
15. Yang H, Wang S, Kang YJ, et al. Long non-coding RNA SNHG1 predicts a poor prognosis and promotes colon cancer tumorigenesis. Oncol Rep. 2018;40(1):261–271.
16. Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi:10.1016/j.biopha.2016.02.036
17. Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8(11):17785–17794. doi:10.18632/oncotarget.14854
18. Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52(1):77–88.
19. Sahu D, Hsu CL, Lin CC, et al. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7(36):58022–58037. doi:10.18632/oncotarget.11158
20. Yu Y, Zhang M, Wang N, et al. Epigenetic silencing of tumor suppressor gene CDKN1A by oncogenic long non-coding RNA SNHG1 in cholangiocarcinoma. Cell Death Dis. 2018;9(7):746. doi:10.1038/s41419-018-0768-6
21. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–1967.
22. Yao J, Zhou B, Zhang J, et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014;35(8):7935–7944. doi:10.1007/s13277-014-1949-2
23. Bian Z, Zhang J, Li M, et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–4819. doi:10.1158/1078-0432.CCR-17-2967
24. Wang P, Peng X, Zhang J, et al. LncRNA-135528 inhibits tumor progression by up-regulating CXCL10 through the JAK/STAT pathway. Apoptosis. 2018;23(11–12):651–666. doi:10.1007/s10495-018-1482-7
25. Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppressing miR-184 expression. Cancer Med. 2017;6(12):2897–2908. doi:10.1002/cam4.1253
26. Wang F, Ji X, Wang J, et al. LncRNA PVT1 enhances proliferation and cisplatin resistance via regulating miR-194-5p/HIF1a axis in oral squamous cell carcinoma. Onco Targets Ther. 2020;13:243–252. doi:10.2147/OTT.S232405
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



