Back to Journals » Cancer Management and Research » Volume 11
Overexpression of HOXC6 promotes cell proliferation and migration via MAPK signaling and predicts a poor prognosis in glioblastoma
Authors Yang P, Kang W, Pan Y, Zhao X, Duan L
Received 25 March 2019
Accepted for publication 10 July 2019
Published 3 September 2019 Volume 2019:11 Pages 8167—8179
DOI https://doi.org/10.2147/CMAR.S209904
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
PengYu Yang, Wei Kang, YaWen Pan, XianJun Zhao, Lei Duan
Department of Neurosurgery, Lanzhou University Second Hospital, Lanzhou University Second Clinical Medical College, Lanzhou University, Lanzhou, People’s Republic of China
Correspondence: Lei Duan
Department of Neurosurgery, Lanzhou University Second Hospital, Lanzhou University Second Clinical Medical College, Lanzhou University, 82 Cuiyingmen, Chengguan District, Lanzhou 730030, People’s Republic of China
Email [email protected]
Background: Homeobox (HOX) genes encode transcription factors that are critical to morphogenesis and cell differentiation. Although the dysregulation of several HOX genes in glioblastoma (GBM) has been reported, little is known about HOXC6 expression in GBM. Therefore, in this study, we investigated the expression levels of the HOXC6 in GBM and explored the regulatory mechanism underlying the role of HOXC6 in GBM progression.
Methods: The ONCOMINE and Oncolnc databases were used to predict the expression level of HOXC6 mRNA and its prognostic value in GBM. The expressions of HOXC6 mRNA in GBM tissues and adjacent brain tissues were detected using qRT-PCR and Western blot. Immunohistochemistry was performed to verify the HOXC6 protein expression in 107 GBM tissues. Kaplan–Meier and Cox analyses were performed to validate the correlation between HOXC6 expression and GBM prognosis. Lentivirus-mediated HOXC6 mRNA overexpression and interference system were established and transfected into U251 and U87 cell lines. CCK-8, colony formation, wound healing and transwell assay were utilized to evaluate the effects of HOXC6 on proliferation and migration of human GBM cells.
Results: High expression of HOXC6 was observed in GBM tissues and GBM cells lines, and it correlated with a decreased overall survival and disease-free survival. Overexpression of HOXC6 promoted the GBM cell proliferation and migration, whereas depletion of HOXC6 reduced GBM cell proliferation and migration. Mechanistic study showed that upregulation of HOXC6 significantly increased the phosphorylation of Jun amino-terminal kinase, ERK and P38, as well as the expression of mitogen-activated protein kinase (MAPK) signaling–related genes, including c-myc, c-jun and p53. Inversely, silencing HOXC6 showed the opposite results.
Conclusion: HOXC6 promoted proliferation and migration of GBM cells via the activation of MAPK pathway.
Keywords: HOXC6, glioblastoma, proliferation, migration, MAPK pathway
Introduction
Glioblastoma (GBM) takes up about 30% of all brain and central nervous system (CNS) tumors and nearly 80% of malignant brain and CNS tumors.1,2 For a better understanding of the etiology and improved personalized therapy for subgroup patients, The Cancer Genome Atlas (TCGA) classified GBMs into four subtypes: proneural, neural, classical and mesenchymal. Each subtype differs greatly in terms of its cellular features, genetic contexts and signaling pathways involved.3 At present, the overall survival (OS) of GBM patients remains rather unsatisfying, with less than 5% of patients surviving for 5 years after the first diagnosis.4–8 Actually, the pathogenic mechanisms contributing to the development and progression of GBM were still not fully elucidated, which may prevent us to develop effective targeted drugs of GBM. Therefore, it is of much urgency to further elucidate the pathogenic mechanisms responsible for the development and progression of GBM.
The homeobox (HOX) gene family members play important roles in the regulation of embryonic development by regulating downstream target genes by their specific DNA binding ability.9 Accumulating evidences show that the abnormal expression of some HOX family genes exists in a variety of tumors, including lung cancer, colorectal cancer, hepatocellular carcinoma, ovarian cancer, pancreatic cancer and breast cancer, and these HOX family members have importantly accelerative effects on the invasive and proliferative abilities of tumor cells.10–15 These previous results indicated that HOX genes may be key regulatory genes for tumors.
Numerous studies have reported that HOXC6, as a member of the HOX gene family, was overexpressed in various cancers, and played a key role in promoting cancer progression, suggesting that HOXC6 was a potential biomarker for diagnostic, prognostic and therapeutic target of cancers.16–18 For instance, Chen et al found that HOXC6 gene significantly promoted the proliferation, invasion and metastasis of gastric cancer cells by upregulation of MMP9, which was associated with the poor prognosis of gastric cancer patients.19 Particularly, a previous study by Guo et al showed that HOXC6 gene expressions in U-118 and U-138 of GBM cell lines were significantly higher than that in normal human astrocytes (NHA), and suppressing HOXC6 expression by introducing corresponding antisense fragments reduced the invasion tendency of U-118 and U-138 cell lines.20 However, little is known about the role of HOXC6 in the development and progression of GBM and the relevant regulatory mechanisms.
In the present study, we found that HOXC6 was overexpressed in GBM tissues and cell lines, and high HOXC6 expression predicted a poor prognosis in GBM patients. Using HOXC6 depletion and overexpression GBM cell models, we demonstrated that HOXC6 promoted tumor cell proliferation and migration, mainly through mitogen-activated protein kinase (MAPK) pathway. We proposed that HOXC6 could be a prognostic biomarker and promising molecular target for GBM.
Materials and methods
Tissue samples and clinical data
A total of 107 tumor specimens and 14 adjacent normal brain tissue (NBT) samples were collected from primary GBM patients who underwent neurosurgery at Lanzhou University Second Hospital. None of the patients received chemotherapy or radiotherapy before surgical tumor resection, and all specimens were obtained during the initial surgery. OS was estimated from the date of diagnosis to the date of either death or last follow-up from 2015 to 2017. Specimens of NBTs were obtained from autopsy material, and to keep these specimens histologically normal, the tissues were acquired within 12 hrs after death. The collection and use of the tissue samples and clinical data were strictly conducted in accordance with the Declaration of Helsinki. Furthermore, all patients provided written informed consent and this study was approved by the Ethics Committee of the Lanzhou University Second Hospital.
Reagents
Commercially available antibodies were as follows: HOXC6 (ab41587, Abcam, Cambridge, UK), GAPDH (60004–1-Ig, Proteintech), Jun amino-terminal kinase (JNK) (#9252, Cell Signaling Technology), Phospho-JNK (#4668, Cell Signaling Technology), p38 (#9212, Cell Signaling Technology), Phospho-p38 (#4511, Cell Signaling Technology), Erk1/2 (#4695, Cell Signaling Technology), Phospho- Erk1/2 (#4370, Cell Signaling Technology), c-Jun (#9165, Cell Signaling Technology) and P53 (#2524, Cell Signaling Technology), c-myc (#13987, Cell Signaling Technology).All other chemical reagents were obtained from Sigma-Aldrich, unless otherwise indicated.
Cell lines and cell culture
Human GBM cell lines, U251, U87, LN229, A172, U118 and NHA, were purchased from the China Academia Sinica Cell Repository (Shanghai, China). U251, U87, LN229, A172, U118 and NHA cells were cultured in DMEM (cat#11965–092,Gibco, USA) or RPMI 1640 (cat#12633012, Gibco, USA) supplemented with 10% FBS (cat#10099-141, Gibco, USA), 50 units/mL penicillin/streptomycin and incubated at 37°C in a humidified incubator (Forma™ 310, Thermo, USA) with 5% CO2/95% air.
Plasmid construction, lentivirus packaging and stable cell lines generation
The complete open reading frame of human HOXC6 was amplified by PCR using the methods according to the study of Zhang X. et al.21 The amplicon was inserted into the HindIII-XbaI site of pcDNA3.1 vector. Short hairpin RNA (shRNA) targeting the HOXC6 was designed as follows: sense a 5ʹ-CCGGGACCTCAATCGCTCAGGATTTCTCGAGAAATCCTGAGCGATTGAGGTCTTTTTG-3ʹ, sense b 5ʹ-AATTCAAAAAGACCTCAATCGCTCA GGATTTCTCGAGAAATCCTGAGCGATTGAGGTC-3ʹ.22 The pcDNA3.1-HOXC6 or shRNA-harboring plasmid and two helper plasmids, pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio, China), were transfected into HEK293T cells using Lipofectamine 2000 (cat#11668019, Life Technologies, USA). Two days after transfection, cell culture media were collected and concentrated. The recombinant lentivirus was stored at −80°C.
RNA extraction, cDNA synthesis and real-time quantitative RT-PCR
Fifty-three samples were collected after surgical resection, snap-frozen and stored in liquid nitrogen until RNA extraction. Total RNA from cryogenically homogenized tumor tissue was isolated using 1.0 mL TRIzol Reagent (cat#15596026, Invitrogen, USA) and purified with the RNeasy Mini Kit (cat#74104, Qiagen, Hilden, Germany). RNase-free DNase (cat#EN0521, ThermoFisher Scientific) was used in order to avoid DNA contamination. Reverse transcription was performed using the first-strand cDNA synthesis Kit (cat#12328032, Invitrogen). qRT-PCR was used to detect the expression levels of HOXC6 using SYBR Premix Ex Taq™ kit (cat#DRR041A, Takara, Japan) following the manufacturer’s introductions and the parameters were as follows: hot start at 95°C for 5 mins, followed by 40 cycles of 95°C for 30 s, 60°C for 45 s and 72°C for 30 mins. The primers for HOXC6 and internal standard GAPDH were designed as follows: HOXC6 forward, 5ʹ-ACAGACCTCAATCGCTCAGGA-3ʹ and reverse, 5ʹ- AGGGGTAAATCTGGATACTGGC-3ʹ (86-bp product); and GAPDH forward, 5ʹ-TCCTCTGACTTCAACAGCGACACC-3ʹ and reverse, 5ʹ-TCTCTCTTCCTCTTGTCGTCTTGG-3ʹ (286-bp product). The relative quantification of the value was determined using the 2−ΔΔCt calculation method and each sample was assayed in triplicate.23
Western blotting
Western blot assay was performed as a standard procedure. Total protein was extracted using radioimmunoprecipitation buffer (cat#2114–100, BioVision) with protease/phosphatase inhibitor cocktail (cat#4693116001, Roche). The protein concentrations were detected by using a bicinchoninic acid protein assay kit (cat#23225, Thermo Fisher Scientific, Inc.). Protein samples (20 μg/well) and dual plus molecular weight ladders (#161–0374, Bio-Rad Laboratories, Hercules, CA) were separated using 8–12% gradient polyacrylamide gel and then transferred onto polyvinylidene difluoride (cat#ISEQ00010, millipore) membranes. The membranes were blocked in Tris-buffered saline (cat#A20120A0211, BioTNT) containing 5% bovine serum albumin (cat#70-A3828-100, MultiSciences) at room temperature for 1 hr and subsequently incubated with the indicated primary antibody at 4°C overnight and then with the secondary antibody at room temperature for 1 hr. Bands were visualized using the enhanced chemiluminescence (cat#32106, ECL, Pierce).24 The density of protein bands was calculated using the ImageJ software (National Institutes of Health, Bethesda, MD, USA, https://imagej.nih.gov/ij/). According to the instruction of ImageJ, we finally calculated the relative density value of each protein band (https://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/).
Immunohistochemistry
Immunohistochemistry was performed as described previously.25 Briefly, paraffin-embedded specimens were cut into 4-mm sections and baked at 65°C for 1 hr. The sections were deparaffined with xylenes and rehydrated, submerged into 0.01 M citrate buffer (pH 6.0) antigen retrieval buffer and microwaved for antigenic retrieval. They were treated with 0.3% H2O2 for 15 mins to block the endogenous peroxidase at RT and then were treated with normal goat serum for 30 mins to reduce the nonspecific binding. Consequently, the sections were incubated with rabbit polyclonal anti-HOXC6 antibody (1:200) overnight at 4°C. After being washed, the sections were treated with anti-rabbit/mouse secondary antibody at room temperature for 1 hr. The tissue sections were immersed in 3-amino-9-ethyl carbazole, counterstained with Mayer’s hematoxylin, dehydrated, and finally mounted in Crystal Mount. The immunohistochemically stained tissue sections were scored independently by two pathologists blinded to the clinical parameters. The intensity and extent of the staining were used as criteria of evaluation. The staining intensity was scored as 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3 (strong staining). Extent of staining was scored as 1 (<25%), 2 (25–50%), 3 (50–75%) and 4 (75–100%), according to the percentages of the positive staining areas in relation to the whole carcinoma area or entire section for the normal samples. Staining index was calculated as the multiplication of staining intensity score and the proportion of HOXC6-positive tumor cells. All scores were subdivided into two categories according to a cutoff value in the study cohort: HOXC6 high expression (>7.5) and low expression (<7.5).
Cell proliferation assay
Cells were plated in 5 replicates in a 96-well plate (3000 cells/well) and cultured in DMEM containing 10% FBS. Then, cells were incubated with 10 uL CCK8 reagent (LOT#GB707, Dojindo Molecular Technologies, Inc., Kumamoto, Japan) for 4 hrs at 37°C. Viable cells were counted every other day by reading the absorbance at 450 nm with a plate reader (ELx800, BioTek, Winooski, USA). Statistical analysis (one-way ANOVA) was performed using the statistical software GraphPad Prism 5. A value of P≤0.05 was considered to be significant. The test was repeated three times.
Colony formation assay
For the colony formation assay, stably infected cells were seeded in 6-well plates at a density of 500 cells/well at an atmosphere of 5% CO2 for 14 days. The growth medium was changed at 3-day intervals. After 14 days, cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet (C6158, Sigma-Aldrich, St. Louis, USA) and dried. Only clearly visible colonies (more than 50 cells) were counted under a light microscope. The test was repeated three times. Only clearly visible colonies (more than 50 cells) were counted under a light microscope. Data were expressed as mean ± standard deviation. Statistical significance was determined using Student’s t-test where p≤0.05 was considered statistically significant.
Wound healing assay
Cells were seeded at in 6-well plates at a density of 4.0×105 cells per well at an atmosphere of 5% CO2. After 1 day, confluent monolayer cells were linearly scratched using a 200-μL pipette chip, one parallel was made per experiment. The scratched region was photographed immediately or 24 hrs after scratching using a microscope equipped with a camera. Five pictures were randomly taken for each experiment. The scratch width from per experiment was calculated, and data were then obtained and expressed as mean ± standard deviation.
Migration assay
A cell migration assay was performed using the Boyden chamber with a gelatin-coated polycarbonate filter with 8-μm pore size (Neuro Probe, Gaithersburg, MD, USA). Cells in serum-free culture medium were added to the upper chamber at a density of 2.0×106 cells (200 μL/well), and the lower chamber was filled with 10% FBS culture medium. After 24 hrs, the nonpenetrated cells were removed using a cotton swab. Cells that had invaded to the back of the membrane of the transwell chamber were stained with 0.1% crystal violet after fixed by 4% formaldehyde. The invasive capacity of cells was defined according to the total number of cells in randomly selected fields by light microscopy. Statistical significance was determined using Student’s t-test where p≤0.05 was considered statistically significant.
ONCOMINE and OncoLnc analysis
ONCOMINE (http://www.oncomine.org) is a collection of cancer microarray databases with an integrated data-mining platform.26 Bredel Brain 2, Lee Brain, Murat Brain, Sun Brain and TCGA Brain datasets were selected because they were established on mRNA levels and contain larger samples (n>50). P-value=1E-4, fold change=1.5, gene rank=top 10% were selected as threshold. OncoLnc (www.oncolnc.org/) is a newly available resource for COX coefficients and linking TCGA survival data to mRNA, miRNA or lncRNA expression. The survival analysis was performed using a cutoff value of median of HOXC6 expression to evaluate the correlation between the expression of HOXC6 and the prognosis of patients with GBM.
Statistical analysis
The SPSS software version 20.0 and GraphPad Prism 5 software were used to perform statistical analyses. The Cox proportional hazards model and Kaplan–Meier analysis were employed for survival analysis, and the significance of variances between two groups was determined by log-rank test. Each experiment was performed three times in triplicate. p<0.05 was considered statistically significant.
Results
HOXC6 is highly expressed in GBM
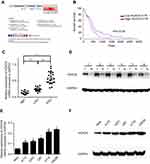
To analyze the transcript levels of HOXC6 in GBM, we adopted an online microarray database ONCOMINE (www.oncomine.org) and found significant overexpression of HOXC6 in the majority of GBM tissues compared with adjacent non-neoplastic controls (Figure 1A). Then, we further assessed the prognostic value of HOXC6 mRNA level in 152 cases of GBM using the OncoLnc database (www.oncolnc.org). The data showed that overexpression of HOXC6 was associated with poorer OS in cancerous tissues compared to paracancerous tissues (Figure 1B). To further explore the relationship between HOXC6 expression levels and GBM, we detected the mRNA expression levels of 14 NBT, 18 samples of low-grade GBM tissue samples (LGG, WHO grade I-II) and 21 samples of high-grade GBM tissue samples (HGG, WHO grade I-II) using qPCR.27 Data confirmed that HOXC6 mRNA levels were more elevated in high-grade GBM tissues than in low-grade GBM tissues, and HOXC6 mRNA levels were found higher in low-grade GBM than in NBTs (Figure 1C, p<0.05). Next, we detected the HOXC6 protein levels in 6 paired primary GBM tissues and the corresponding adjacent normal tissues using Western blotting. Results showed that HOXC6 protein levels (6/6,100%) were increased in GBM tissues compared with NBTs (Figure 1D). To further explore the potential role of HOXC6 in GBM tumorigenesis, we detected the mRNA expression and protein expression of HOXC6 in 1 normal brain and 5 different GBM cell lines using Western blotting. Compared with the normal brain cell lines NHA, HOXC6 was overexpressed in all the detected GBM cell lines (Figure 1E and F). Taken together, these data indicated that HOXC6 expression was increased in GBM tissues and cells.
Upregulation of HOXC6 is associated with poor prognosis in GBM
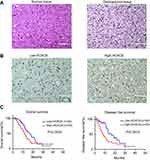
To further investigate the HOCX6 expression in fresh GBM tissues, we employed H&E staining and IHC staining with 14 NBT samples and 107 GBM samples from Lanzhou University Second Hospital. As shown in Figure 2A, NBT and glioma tissue were observed and differentiated by H&E staining assay. Then, we identified that HOCX6 expression was specifically detected in the cytoplasm of GBM cells from the cancer tissues in the majority of GBM samples (90/107 cases, 84.1%), while others presented negative expression (17/107 cases, 15.9%). No signals were found in all the corresponding adjacent normal tissues or normal tissue (Figure 2B). According to the IHC results, 54 cases were defined as low HOCX6 expression, while the other 53 cases were identified as high expression. According to Kaplan–Meier survival analysis, patients with higher HOXC6 expression predicted a decreased OS (p=0.0243) and disease-free survival (p=0.0013, Figure 2C). These data indicated that HOXC6 could be a prognostic marker for GBM patients.
HOXC6 promotes proliferation and clonogenicity of GBM cells
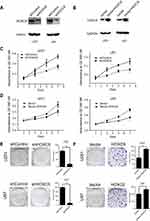
To address the potential role of HOXC6 in GBM progression, HOXC6 knockdown and overexpression of GBM cell models were constructed in U251 and U87 cells with lentivirus infection method. High efficiency of knockdown and overexpressing cell models was demonstrated to be successfully established (Figure 3A and B). CCK-8 assay was then performed to assess cell viability at the indicated times. Data showed that the inhibition of HOXC6 markedly declined the GBM cells’ viability (Figure 3C). On the contrary, cellular proliferation ability greatly increased after HOXC6 overexpression (Figure 3D). Colony formation assay showed that, compared with the shNC cells, both the size and number of shHOXC6 cell group were dramatically decreased (Figure 3E). On the other hand, the size and number were significantly increased in HOXC6-overexpressing cells (Figure 3F). Taken together, these results indicated that HOXC6 played an important role in GBM proliferation and clonogenicity.
HOXC6 promotes GBM cells migration
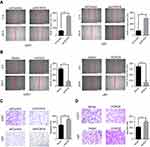
Wound healing and transwell assay were performed to detect the effect of HOXC6 on GBM cell migration. As shown in Figure 4A, after 24 hrs, the width of the wound in shHOXC6 group was markedly wider than that in shControl group. On the contrary, the width of the wound greatly decreased after HOXC6 overexpression (Figure 4B). Next, we examined the role of HOXC6 in GBM cell migration using transwell chambers. The number of migrating HOXC6 knockdown GBM cells was significantly reduced compared to shControl group (Figure 4C). Conversely, we found that the number of HOXC6-overexpressing cells was significantly increased compared to the empty vector group (Figure 4D). These results indicated that HOXC6 could facilitate to GBM cells migration.
HOXC6 is associated with transcriptional activity of the MAPK pathway
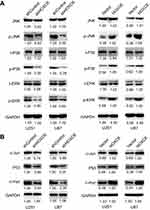
Recently, Zhang F. et al have revealed that HOXC6 gene silencing may inhibit EMT event and cell viability in cervical carcinoma through the inhibition of TGF-β/smad signaling pathway.28 Moreover, substantial differences in gene expression between TGF-beta1 sensitive and insensitive cell lines were observed involving genes in MAPK pathways.29 Besides, Guo C. et al have reported that HOXC10 upregulation significantly increased mRNA and protein expression of MAPK signaling-related genes.30 Because HOXC6 and HOXC10 belong to the same gene family and share the same conserved region, we speculated that expression levels of HOXC6 may be involved in the MAPK pathway. Western blotting results showed that levels of phosphorylation of P38, JNK and ERK were downregulated in the protein lysate of shHOXC6 cells group, while the total amount of JNK, P38 and ERK remained unchanged (Figure 5A). More importantly, as important downstream molecules of MAPK pathway, expression of MYC proto-oncogene (c-myc), Jun proto-oncogene, AP-1 transcription factor subunit (c-Jun) and P53 were downregulated. Similarly, results in HOXC6-overexpressing cell lines showed that total amount of JNK, P38 and ERK remained unchanged, protein levels of phosphorylation of P38, JNK, and ERK were upregulated, expression of downstream molecules of MAPK pathway, c-myc, c-Jun and P53 were upregulated (Figure 5B). These results indicated that HOXC6 may contribute to proliferation and migration of GBM cells by regulating the MAPK pathway.
Discussion
GBM, the most frequent type of primary brain tumor, is one of the most aggressive and deadly human cancers.31–33 Regardless of the huge advance in sGBM therapy over the past several decades, GBM patients’ median survival rate has not significantly improved.34–36 A large number of evidence suggest that abnormal expression of certain genes is associated with tumor development and progression.37–42 Therefore, growing attention has been paid to explore the roles of aberrant expression of certain genes in tumor progression, such as proliferation, metastasis, stemness and drug resistance, which may help to develop molecular target therapy.43,44 However, up to date, only a few effective target therapies are available for GBM patients. Therefore, there is still a great need to identify new molecules related to GBM progression. In our study, we demonstrated that higher expression of HOXC6 significantly correlated with poorer prognosis in patients with gliomas. In addition, HOXC6 could contribute to the GBM cell proliferation and migration mainly through activating MAPK signaling pathway.
HOXC6 gene is one of the homeoprotein transcription factors involved in regulating embryonic development and several cellular processes such as differentiation and morphogenesis.28 The function and underlying molecular mechanisms of HOXC6 in cancer progression are multifaceted and remain elusive. In several types of tumors, HOXC6 is upregulated and serves as a tumor activator. For instance, overexpressed HOXC6 is frequently observed in esophageal squamous cell carcinoma compared to paracancerous tissues and relates to poorer OS.9 Overexpressed HOXC6 is closely linked to poor survival of patients with gastric cancer and contributes to gastric metastasis.17 Besides, it was also reported that HOXC6 was upregulated in colorectal cancer.22 However, Tait DL et al indicated that HOXC6 was downregulated and might serve as a tumor suppressor in serous ovarian cancer.45 Consistent with most of the previous studies, in our current study, we found that HOXC6 may serve as an oncogene in GBM tumorigenesis by ONCOMINE and OncoLnc database analysis. Meanwhile, our results from RT-PCR and Western blot assays confirmed the upregulation of HOXC6 in GBM clinical tissue samples. In addition, HOXC6 was also obviously upregulated in U87, U251, U172, U118 and LN229 cell lines in comparison with normal NHA cells. Furthermore, we demonstrated that HOXC6 exhibited a stronger immunostaining in the cytoplasm of GBM cells compared with benign tissues and its increased expression correlated with poor survival. These results indicated that HOXC6 was a highly expressed tumor-associated gene and could be exploited as a prognostic marker for GBM patients.
Unlimited proliferation and metastasis, as key hallmarks of cancers, are the two main causes of tumor progression and poor prognosis.46–49 Hence, in this study, we focused on investigating the effects of HOXC6 on the biological behaviors of GBM cells lines and found that overexpression of HOXC6 in U251 and U87 cells promoted their proliferation and migration whereas knockdown of HOXC6 showed the opposite results. The generic MAPK signaling pathway is shared by three main distinct cascades, including the ERKs (ERK1/2), JNKs (JNK1/2/3) and p38-MAPK, and has been reported to be associated with cancer cell proliferation, differentiation, migration, senescence and apoptosis.50,51 A study by Guo C et al52 suggested that HOXC10 upregulation significantly increased the mRNA and protein expression of MAPK signaling–-related genes and their phosphorylation. Because HOXC6 and HOXC10 belong to the same gene family and share the same conserved region, we speculated that HOXC6 may be involved in regulating the MAPK pathway. Following this, we tried to explore whether HOXC6 exerted its effect on biological functions of GBM cells lines through the MAPK pathway. Our results showed that overexpression of HOXC6 increased the phosphorylation of JNK/ERK/P38 and their downstream genes (c-myc, P53 and c-Jun), while interfering HOXC6 expression showed the inverse effects. Taken together, HOXC6 may promote the proliferation and migration of GBM cell lines by activating the MAPK signaling pathway.
The present study has some limitations. First, the population of the enrolled patients was relatively small, which might result in a degree of bias. Therefore, more studies with a large sample are needed to validate our findings. Second, many factors, such as some key patient baselines (age, tumor stage, tumor differentiation, adjuvant therapy, etc.) and the other aberrant expression of oncogenes are considered to affect the survival of cancer patients. However, in this study, we did not incorporate these factors and HOXC6 expression status into the Cox proportional hazards regression model for multivariate survival analysis. Therefore, to accurately assess the prognostic value of HOXC6, in future study, we and the other researchers should collect these clinical data for performing more comprehensive prognosis analysis. Third, animal experiments are needed to validate HOXC6’s function in vivo, and further investigations are awaited to explain the exact molecular mechanism behind the association of HOXC6 expression with MAPK pathway.
Conclusion
Our study showed that HOXC6 promoted the proliferation and migration of GBM cell via MAPK pathway in vitro and was closely associated with poor GBM prognosis, suggesting that may be a diagnostic, prognostic and therapeutic biomarker for GBM.
Acknowledgment
This work was supported by grant from Gansu Province Health Industry Research Project (GSWSKY2018-01).
Disclosure
The authors declared no conflicts of interest in this work.
References
1. Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi:10.1038/nrdp.2015.17
2. Ostrom QT, Haley G, Paul F, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;18(1):204.
3. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi:10.1016/j.ccr.2009.12.020
4. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro-Oncology. 2014;16:896–913. doi:10.1093/neuonc/nou087
5. Hatoum A, Mohammed R, Zakieh O. The unique invasiveness of glioblastoma and possible drug targets on extracellular matrix. Cancer Manag Res. 2019;11:1843–1855. doi:10.2147/CMAR.S186142
6. Peng H, Li Z, Fu J, Zhou R. Growth and differentiation factor 15 regulates PD-L1 expression in glioblastoma. Cancer Manag Res. 2019;11:2653–2661. doi:10.2147/CMAR.S192095
7. Syed M, Liermann J, Verma V, et al. Survival and recurrence patterns of multifocal glioblastoma after radiation therapy. Cancer Manag Res. 2018;10:4229–4235. doi:10.2147/CMAR.S165956
8. Ren Z, Liang J, Zhang P, Chen J, Wen J. Inhibition of human glioblastoma cell invasion involves PION@E6 mediated autophagy process. Cancer Manag Res. 2019;11:2643–2652. doi:10.2147/CMAR.S200151
9. Du YB, Dong B, Shen LY, Yan WP. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res. 2014;188:442–450. doi:10.1016/j.jss.2014.01.017
10. Liu H, Zhang M, Xu S, et al. HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer. Oncogenesis. 2018;7(2):1. doi:10.1038/s41389-017-0016-4
11. Watanabe Y, Saito M, Saito K, et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol Lett. 2018;15:2756–2762. doi:10.3892/ol.2017.7650
12. Quagliata L, Quintavalle C, Lanzafame M, et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab Invest. 2017;98(1):95. doi:10.1038/labinvest.2017.107
13. Li BIN, Jin H, Yu Y, Gu CHAO, Zhou X, Zhao N. 1042 human homeobox gene (HOX) A10 is overexpressed in human ovarian clear cell adenocarcinoma and correlates with poor survival. Int J Gynecol Cancer. 2010;7:99.
14. Tsuboi M, Taniuchi K, Shimizu T, Saito M, Saibara T. The transcription factor HOXB7 regulates ERK kinase activity and thereby stimulates the motility and invasiveness of pancreatic cancer cells. J Biol Chem. 2017;292:17681. doi:10.1074/jbc.M116.772780
15. Ji-Yeon L, Ho H, Hyo Jung Y, et al. HOXB5 promotes the proliferation and invasion of breast cancer cells. Int J Biol Sci. 2015;11:701–711. doi:10.7150/ijbs.11431
16. Ya-Bing D, Bin D, Lu-Yan S, et al. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res. 2014;188:442–450. doi:10.1016/j.jss.2014.01.017
17. Zhang Q, Jin XS, Yang ZY, Wei M, Liu BY, Gu QL. Upregulated Hoxc6 expression is associated with poor survival in gastric cancer patients. Neoplasma. 2013;60:439–445. doi:10.4149/neo_2013_057
18. Chang SL, Chan TC, Chen TJ, Lee SW, Lin LC, Win KT. HOXC6 overexpression is associated with Ki-67 expression and poor survival in NPC patients. J Cancer. 2017;8:1647–1654. doi:10.7150/jca.18893
19. Chen SW, Zhang Q, Xu ZF, et al. HOXC6 promotes gastric cancer cell invasion by upregulating the expression of MMP9. Mol Med Rep. 2016;14:3261. doi:10.3892/mmr.2016.5640
20. Guo YB, Shao YM, Chen J, et al. Effect of overexpression of HOX genes on its invasive tendency in cerebral glioma. Oncol Lett. 2016;11:75. doi:10.3892/ol.2016.4099
21. Zhang X, Hamada JA, Takahashi Y, Murai T, Tada M, Moriuchi T. HOXC6 and HOXC11 increase transcription of S100beta gene in BrdU-induced in vitro differentiation of GOTO neuroblastoma cells into schwannian cells. J Cell Mol Med. 2010;11:299–306. doi:10.1111/j.1582-4934.2007.00020.x
22. Ji M, Feng Q, He G, et al. Silencing homeobox C6 inhibits colorectal cancer cell proliferation. Oncotarget. 2016;7:29216–29227. doi:10.18632/oncotarget.8703
23. Skiriutė D, Steponaitis G, Vaitkienė P, et al. Glioma malignancy-dependent NDRG2 gene methylation and downregulation correlates with poor patient outcome. J Cancer. 2014;5:446–456. doi:10.7150/jca.9140
24. Huang Y, Wang H, Lian Y, et al. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis. 2018;9:141. doi:10.1038/s41419-018-1111-y
25. Lian YF, Huang YL, Wang JL, et al. Anillin is required for tumor growth and regulated by miR-15a/miR-16-1 in HBV-related hepatocellular carcinoma. Aging (Albany NY). 2018;10:1884–1901. doi:10.18632/aging.101510
26. Rhodes DR, Shanker KS, Vasudeva M, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi:10.1593/neo.07112
27. Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120:567–584. doi:10.1007/s00401-010-0736-4
28. Zhang F, Ren CC, Liu L, Chen YN, Yang L, Zhang XA. HOXC6 gene silencing inhibits epithelial-mesenchymal transition and cell viability through the TGF-β/smad signaling pathway in cervical carcinoma cells. Cancer Cell Int. 2018;18(1):204. doi:10.1186/s12935-018-0680-2
29. Kloth JN, Gert Jan F, Jan O, et al. Substantial changes in gene expression of Wnt, MAPK and TNFalpha pathways induced by TGF-beta1 in cervical cancer cell lines. Carcinogenesis. 2005;26:1493–1502. doi:10.1093/carcin/bgi110
30. Chen J, Sheng X, Deng G. HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. 中国癌症研究:英文版. 2017;29:572–580.
31. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi:10.1056/NEJMra0708126
32. Wang J, Liang H, Sun M, et al. Delta-6-desaturase inhibitor enhances radiation therapy in glioblastoma in vitro and in vivo. Cancer Manag Res. 2018;10:6779–6790. doi:10.2147/CMAR.S185601
33. Wang Z, Yu G, Liu Z, et al. Paeoniflorin inhibits glioblastoma growth in vivo and in vitro: a role for the Triad3A-dependent ubiquitin proteasome pathway in TLR4 degradation. Cancer Manag Res. 2018;10:887–897. doi:10.2147/CMAR.S160292
34. Shota T, Louis DN, Curry WT, Batchelor TT, Jorg D. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14–26. doi:10.1038/nrclinonc.2012.204
35. Chen Z, Xu N, Zhao C, Xue T, Wu X, Wang Z. Bevacizumab combined with chemotherapy vs single-agent therapy in recurrent glioblastoma: evidence from randomized controlled trials. Cancer Manag Res. 2018;10:2193–2205. doi:10.2147/CMAR.S173323
36. Poulsen HS, Urup T, Michaelsen SR, Staberg M, Villingshoj M, Lassen U. The impact of bevacizumab treatment on survival and quality of life in newly diagnosed glioblastoma patients. Cancer Manag Res. 2014;6:373–387. doi:10.2147/CMAR.S39306
37. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi:10.1016/j.cell.2011.02.013
38. Wang YP, Huang LY, Sun WM, et al. Insulin receptor tyrosine kinase substrate activates EGFR/ERK signalling pathway and promotes cell proliferation of hepatocellular carcinoma. Cancer Lett. 2013;337:96–106. doi:10.1016/j.canlet.2013.05.019
39. Chen H, Zhu HR, Yu XN, et al. Overexpressed pepsinogen C is associated with poor prognosis in human hepatocellular carcinoma: a tissue microarray study. Cancer Manag Res. 2019;11:2927–2934. doi:10.2147/CMAR.S192241
40. Changyong E, Yang J, Li H, Li C. LncRNA LOC105372579 promotes proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma via activating miR-4316/FOXP4 signaling. Cancer Manag Res. 2019;11:2871–2879. doi:10.2147/CMAR.S197979
41. Li Y, Jiao Y, Fu Z, Luo Z, Su J, Li Y. High miR-454-3p expression predicts poor prognosis in hepatocellular carcinoma. Cancer Manag Res. 2019;11:2795–2802. doi:10.2147/CMAR.S196655
42. Zhu L, Pan R, Zhou D, Ye G, Tan W. BCL11A enhances stemness and promotes progression by activating Wnt/beta-catenin signaling in breast cancer. Cancer Manag Res. 2019;11:2997–3007. doi:10.2147/CMAR.S199368
43. Philippe B, Philippe A, Ahmad A. New generations of targeted therapies fighting the resistance in solid tumors. Curr Opin Oncol. 2015;27:243. doi:10.1097/CCO.0000000000000175
44. Christian R, Giuseppe B, Giovanni S, et al. The role of targeted therapy for gastrointestinal tumors. Expert Rev Gastroenterol Hepatol. 2014;8:875–885. doi:10.1586/17474124.2014.922870
45. Tait DL, Bahrani-Mostafavi Z, Vestal CG, Richardson C, Mostafavi MT. Downregulation of HOXC6 in serous ovarian cancer. Cancer Invest. 2015;33:303–311. doi:10.3109/07357907.2015.1041641
46. Kim S. New and emerging factors in tumorigenesis: an overview. Cancer Manag Res. 2015;7:225–239. doi:10.2147/CMAR.S47797
47. Shahruzaman SH, Fakurazi S, Maniam S. Targeting energy metabolism to eliminate cancer cells. Cancer Manag Res. 2018;10:2325–2335. doi:10.2147/CMAR.S167424
48. Tay RY, Wong R, Hawkes EA. Treatment of metastatic colorectal cancer: focus on panitumumab. Cancer Manag Res. 2015;7:189–198. doi:10.2147/CMAR.S71821
49. Valdespino V, Valdespino PM. Potential of epigenetic therapies in the management of solid tumors. Cancer Manag Res. 2015;7:241–251. doi:10.2147/CMAR.S70358
50. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi:10.3109/10799893.2015.1030412
51. Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89:867–882. doi:10.1007/s00204-015-1472-2
52. Guo C, Hou J, Ao S, Deng X, Lyu G. HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chin J Cancer Res. 2017;29:572–580. doi:10.21147/j.issn.1000-9604.2017.06.12
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.