Back to Journals » OncoTargets and Therapy » Volume 11
Overexpression of Beclin1 inhibits proliferation and promotes apoptosis of human laryngeal squamous carcinoma cell Hep-2
Received 11 August 2017
Accepted for publication 28 January 2018
Published 4 July 2018 Volume 2018:11 Pages 3827—3833
DOI https://doi.org/10.2147/OTT.S148869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Jianmin Xu
Baoluo Wan, Yanzi Zang, Lin Wang
Department of Otorhinolaryngology, Henan Province People’s Hospital, Zhengzhou, Henan Province, China
Objective: Beclin1 was previously found to be downregulated in human laryngeal cancer (LC) tissues, and it results in poor prognosis. This study aimed to further confirm the antitumor effects of Beclin1 in LC cell line Hep-2.
Materials and methods: Beclin 1 was overexpressed in Hep-2 cells using liposomal transfection and confirmed using reverse transcription polymerase chain reaction and Western blotting. Then, cell proliferation and apoptosis were determined in control (untransfected), empty vector transfected, and Beclin1 overexpressed groups using MTT and flow cytometry procedure, respectively.
Results: The expression of the Beclin1 gene in Hep-2 cells was significantly increased after vector transfection compared with control (1.173±0.046 vs 0.453±0.016, P<0.01) and empty vector (1.173±0.046 vs 0.440±0.021, P<0.01). Overexpression of Beclin1 inhibited proliferation at 4 days (0.619±0.051 vs 0.891±0.081 and 0.619±0.051 vs 0.878±0.105, P<0.01), 5 days (0.684±0.078 vs 1.127±0.094 and 0.684±0.078 vs 1.162±0.117, P<0.01), and 6 days (0.725±0.069 vs 1.168±0.103 and 0.725±0.069 vs 1.194±0.097, P<0.01) and promoted apoptosis (14.48%±1.42% vs 4.07%±0.66% and 14.48%±1.42% vs 4.39%±0.80%, P<0.01) in Hep-2 cells in comparison with the control and empty vector groups, respectively.
Conclusion: Beclin1 may be an underlying target for the treatment of LC. This study has provided some experimental basis for the gene therapy of LC.
Keywords: laryngeal cancer, antitumor effect, cell proliferation, cell apoptosis
Introduction
Laryngeal cancer (LC) is a common head and neck cancer characterized by hoarseness, sore throat, and persistent cough, and the incidence is increasing.1,2 Radiotherapy and surgery are the most common therapies used recently for the treatment of LC.3,4 However, surgery is always associated with the loss of language function, and patients undergoing radiotherapy treatment are prone to relapse. Therefore, it is urgent to explore a new therapy for LC.
Autophagy is a highly conserved catabolic process involving the lysosomal transport of proteins and organelles.5 Disorders of this system are associated with many human diseases and the clearance of cancer cell carcasses.6,7 Beclin1 is the encoding gene of autophagy-associated protein Beclin 1, which is a component of the PI3K complex involved in the vesicular transport. Previous studies have shown the important role of Beclin1 in cancers such as breast cancer,8 CaSki cervical cancer,9 lung cancer,10 MIA PaCa-2 pancreatic cancer,11 and tongue squamous cell carcinoma.12 A recent study has also revealed that the expression of Beclin1 is significantly decreased in laryngeal squamous cell carcinoma than that in non-tumor tissues and is obviously associated with the lymph node metastasis and poor survival rate, suggesting that Beclin 1 may be a potential target for the treatment of LC.13 However, the role of Beclin1 in cells was not verified by experiment.
To further reveal its biofunction, Beclin 1 was overexpressed in a human LC cell line hep-2, and the effects of Beclin 1 on cell proliferation and apoptosis were determined in this study, so that it could provide a theoretical basis for the mechanism of Beclin 1 in LC.
Materials and methods
Construction of pIRES2-AcGFP-Beclin1 expression vector and cell transfection
The amplification of Beclin1 gene was performed by polymerase chain reaction (PCR) using the full-length Beclin1 cDNA clone (pCMVsPORT6-Beclin1 from Funeng Genes Co., Ltd, Guangzhou, China) as the template, and then electrophoretic detection was performed. The amplified product was digested with SacI and EcoRI (Promega Corporation, Fitchburg, WI, USA) and purified using the DNA gel recovery kit (Dongsheng Biotech Co., Ltd., Guangzhou, China) according to the manufacturer’s instructions. The target gene Beclin1 was ligated to the expression vector pIRES2-AcGFP (Clontech Laboratories, Inc., Mountain View, CA, USA) and transformed into competent DH5α, and then the cloned recombinants were screened. The recombinant plasmid pIRES2-AcGFP-Beclin1 was extracted with a small-scale plasmid extraction kit from Shanghai Labaide. Finally, the extracted plasmid was transfected into human laryngeal carcinoma Hep-2 cell line (Nanjing KeyGen Biotech, Nanjing, Jiangsu, China) using Lipo2000 according to the manufacturer’s protocol. Hep-2 cells transfected with pIRES2-AcGFP-Beclin1 were considered as the Beclin1 group and Hep-2 cells transfected with pIRES2-AcGFP as the empty vector group, while the control group consisted of the Hep-2 cells not transfected with pIRES2-AcGFP.
Real-time reverse transcription polymerase chain reaction (RT-PCR)
Primers of genes were synthesized in Shanghai Biological Engineering Co., Ltd (Shanghai, China) as follows: Beclin1, forward sequence: 5′ACAGAGCTCATGGAAGGGTCTAAGACGTCC-3′ and reverse sequence: 5′-TACGAATTCTCATTTGTTATAAAATTGTG-3′; and GAPDH, forward sequence: 5′-ACCACAGTCCATGCCATCAC-3′ and reverse sequence: 5′-TCCACCACCCTGTTGCTGTA-3′. Total mRNA was extracted from cells using an mRNA rapid extraction kit (Shanghai Jie Rui Bioengineering Co., Ltd). Then, mRNA was reverse transcribed into cDNA in 30 μL transcription reaction system by AMV with the universal OLIGO hexamer as the reverse transcription primer. The PCR program for Beclin1 and GAPDH was designed as follows: 94°C for 3 min, 35 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 60 s, and finally extension at 72°C for 7 min. The PCR products of Beclin1 gene and internal controls were detected by electrophoresis in agarose gel.
Western blotting
After culturing for 48 h, the cell culture medium was discarded and washed twice with PBS. Then, the cells were collected and lysed with WIP cell lysate (Beijing Boosen Biotechnology Company, Beijing, China) at 4°C for 15 min. Following this, lysates were centrifuged at 4°C for 10 min, and the concentration of supernatant of each protein sample was determined using the bicinchoninic acid protein concentration assay kit (Thermo Fisher Scientific, Waltham, MA, USA). The collected protein samples were boiled with sodium dodecyl sulfate polyacrylamide gel electrophoresis protein loading buffer (5×) for 3–5 min and then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Thereafter, the gel was equilibrated in transfer buffer for 15 min and transferred using Bio-Rad’s standard wet transfer device. After the transfer, the protein membrane was put into the Western washing solution for 1–2 min and blocked with Western blocking solution at room temperature for 1 h. Following this, the membrane was incubated with a mouse anti-human Beclin1 antibody (1:200; Santa Cruz Biotechnology Inc., Dallas, TX, USA) and incubated at room temperature for 1 h. Then, the membrane was incubated with a horseradish peroxidase-conjugated anti-mouse IgG antibody (1:3,000; Santa Cruz Biotechnology Inc.) for 1 h at room temperature. Subsequently, the membrane was rinsed four times with the Western wash solution for 5–10 min and visualized using the enhanced chemiluminescence method. The images were saved using the imaging scanning analysis system (Syngene, Cambridge, UK).
MTT assay
The culture medium was discarded, and the cells were washed twice with PBS. Then, 80 μL of fresh RPIM-1640 medium and 20 μL of 0.5% MTT solution (dissolved in PBS) were added to each well and cultured with cells for 5 h. The supernatant was discarded and 150 μL of dimethyl sulfoxide was added to each well shaking for 10 min at dark condition. The OD of the absorbance at 570 nm was measured by enzyme-linked immunosorbent assay. The growth curve was plotted based on the average of the OD values.
Flow cytometry analysis
In this assay, sequence of Beclin1 was inserted into pcDNA3.1 vector and transfected into hep-2 cells using Lipo2000. Then, the cells were divided into control group (treated only with Lipo2000), empty vector group (transfected with vector), and Beclin1 groups. After transfecting for 48 h, the cells were harvested in 10 mL centrifuge tube and washed with PBS for two times after cooling. Then, the supernatant was discarded and the cells’ concentration was adjusted to 1×106/mL with PBS. After centrifuging for 5 min at 1,000 rpm, the supernatant was removed and the cells were washed once with the incubation buffer followed by centrifugation for 5 min at 1,000 rpm. Subsequently, the cells were resuspended in 100 μL of the labeling solution and incubated for 15 min at room temperature in the dark, and then centrifuged for 5 min at 1,000 rpm. The precipitated cells were collected and washed once with the incubation buffer. The cells were incubated with the fluorescent solution at 4°C for 20 min in dark. The apoptotic rate was detected using flow cytometry with the wavelength of the excitation light at 488 nm. The filter with a wavelength >560 nm was used to detect the propidium iodide.
Statistical analyses
All statistical analyses were performed with SPSS18.0 software (SPSS Inc., Chicago, IL, USA). The results were analyzed by one-way analysis of variance and were expressed as mean ± SD. P<0.01 was considered statistically significant.
Results
Construction of pIRES2-AcGFP-Beclin1 vector
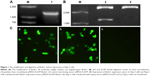
In order to identify the extraction of target genes, Beclin1 gene was amplified using specific primers. The results of electrophoresis showed a bright band at 1,353 bp, which was consistent with the full length of Beclin1 gene (Figure 1A). To confirm the recombination of target Beclin1 gene and the expression vector pIRES2-AcGFP, the vector was digested with SacI and EcoRI and detected by agarose gel electrophoresis. The results showed that there was a bright band at about 1,000 bp of the Beclin1 group (Figure 1B). Because the expression vector had a green fluorescent protein AcGFP marker, the transfection efficacy was also determined by the color of cells under a microscope (Olympus Corporation, Tokyo, Japan) at a wavelength of 450–490 nm. As a result, green-colored cells were identified in both Beclin1 group and empty vector group (Figure 1C), indicating that gene transfection screening was successful.
Expression of Beclin1 mRNA in Hep-2 cells
RT-PCR was used to detect the relative mRNA expression of Beclin1 in Hep-2 cell line. The results showed that the relative mRNA expression of Beclin1 was significantly higher in the Beclin1 group than in the empty vector group (1.173±0.046 vs 0.453±0.016, P<0.01) and the control group (1.173±0.046 vs 0.440±0.021, P<0.01). The results showed that the expression of Beclin1 mRNA in Hep-2 cells could be enhanced by transfection of Beclin1 into Hep-2 cells (Figure 2A and B).
Protein expression of Beclin1 in Hep-2 cells
Western blot was performed to verify whether Beclin1 protein can be successfully expressed in transfected cells. The result showed an obvious lighter imprinted band at ~60 kDa in the Beclin1 group than in the control and empty vector groups (Figure 2C). This finding indicated that the Beclin1 expression vector pIRES2-AcGFP-Beclin1 was successfully transfected into Hep-2 cells.
Overexpression of Beclin1 significantly inhibits the growth of Hep-2 cells
To understand the effect of autophagy gene Beclin1 on the proliferation of Hep-2 cells, the OD values of these three groups were measured by MTT assay. At 4 days, the OD values of Beclin1 group, empty vector group, and control group were 0.619±0.051, 0.891±0.081, 0.878±0.105, respectively. Besides, the OD values of Beclin1 group, empty vector group, and control group were 0.684±0.078, 1.127±0.094, 1.162±0.117, respectively, at 5 days. Moreover, the OD value of Beclin1 group at 6 days was also obviously lower than those of empty vector group and control group (0.725±0.069 vs 1.168±0.103, 0.725±0.069 vs 1.194±0.097, respectively). The results showed that the proliferation of cells in the Beclin1 group was significantly attenuated compared with that in the empty vector and control groups from 4 to 6 days (P<0.01). However, there was no significant difference among these three groups before 3 days (Figure 3A). Then, the cell growth at different culture time points was observed under a microscope. Microscopic observation showed that there were no significant differences in cell growth among these three groups within 3 days (Figure 3B). However, after 3 days of cell culture, the growth rate of the Beclin1 group was significantly inhibited compared with those of empty vector and control groups, but no difference was identified between the control and empty vector groups (P>0.05). At the same time, the numbers of aging (cell morphology rounded, intracellular granules) and dead (cell roundness, floating) cells were obviously increased in the Beclin1 group compared with the control and empty vector groups (Figure 3C).
Overexpression of Beclin1 increases the apoptosis of Hep-2 cell lines
To determine the autophagy effect of Beclin1 on apoptosis of human laryngeal carcinoma Hep-2 cell lines, the apoptotic cells were stained with annexin V-fluorescein isothiocyanate, and the apoptosis rate was determined by flow cytometry. The apoptosis rate of the Beclin1 group was significantly higher than those of the empty vector group (14.48%±1.42% vs 4.07%±0.66%, P<0.01) and the control group (14.48%±1.42% vs 4.39%±0.80%, P<0.01; Figure 4). This evidence indicated that the overexpression of Beclin1 in Hep-2 cells could increase the apoptosis of Hep-2 cells.
Discussion
Although studies have shown that low expression of Beclin1 has a close relationship to poor prognosis of LC, in-depth investigation of the mechanism is rarely reported till now. Therefore, this study aimed to demonstrate the central role of Beclin1 in LC in vitro. Our results showed that overexpression of Beclin1 significantly suppressed cell proliferation, but obviously increased cell apoptosis in Hep-2 cells, which were consistent with previous studies.12,14–16
Autophagy is closely related to the development of cancer. However, the role of autophagy in cancer remains controversial; for example, a study has reported enhancement of autophagy in cancer cell metastasis,17 but the inhibition is suggested by another study.16 There are many claims about the mechanism of Beclin1 in cancers. For example, Beclin1 regulates the cell growth in breast and lung cancers,18,19 inhibits cell proliferation, invasion, and migration in cervical cancer and tongue squamous cell carcinoma,12,16 and induces apoptosis in lung and pancreatic cancers.10,11 Although several studies have reported Beclin1 in many cancers,19,20 the investigation of Beclin1 in LC is rarely reported.
Autophagy and apoptosis are crucial cellular housekeeping and tissue survival mechanisms, and Beclin1 is a platform protein that mediates the crosstalk between apoptosis and autophagy.21 A previous study demonstrated that Beclin1 regulates the autophagic activity by complexing with the type III PI3K. RNase L cleavage products promote autophagy switch to apoptosis by caspase-mediated cleavage of Beclin1.22 Liu et al have documented that CHOP mediates ASPP2-induced autophagic apoptosis by releasing Bcl-2 from Beclin1 and promoting nuclear location of Bcl-2 in hepatoma.23 These findings indicated that Beclin1 plays a crucial role in regulating autophagy and apoptosis. Caspase-mediated cleavage of Beclin1 inactivates Beclin1-induced autophagy and promotes apoptosis by increasing the release of proapoptotic factors from the mitochondria.24 Autophagy inhibits apoptosis by phagocytosing apoptotic pro-caspases such as caspase-8.25 Beclin1 was previously reported as an important gene in the prognosis of LC.13 In this study, we found that overexpression of Beclin1 increased the apoptosis of Hep-2 cells in vitro. Therefore, we deduced that abnormal overexpressed Beclin1 in Hep-2 might trigger some underlying mechanism that could cleave Beclin1 to promote the apoptosis of Hep-2 cell. In fact, autophagy and apoptosis exhibit certain inhibition and have common stimuli and signaling pathways.26,27 As a mediator between these two cell activities, Beclin 1 performs a double-sided regulation of autophagy. On the one hand, autophagy renders the cells chemically resistant by contributing to glycolysis, oxidative metabolism, and cell adhesion–dependent growth, and thereby helps in cell proliferation.28 On the contrary, Beclin1 plays an antiapoptotic role in many stresses interacting with Bcl-2.29,30 For example, caspase-3 and caspase-8 could co-work to cleave Beclin1 into C-terminal orienting to mitochondria to inhibit cells autophagy and N-terminal.26,27,31 In this study, the growth of Hep-2 cells was not significantly changed within 3 days, while the proliferation was significantly decreased in 4–6 days after transfection of Beclin1. Considering this, we suspect that promotion of apoptosis by Beclin 1 might be the potential explanation for the decrease in proliferation of Hep-2 cells. Taken together, Beclin1 might perform a critical role in the survival of Hep-2 cells and can be used as a target of gene therapy for LC.
There were some limitations in this study. For example, although Beclin1 gene significantly inhibited cell proliferation and increased apoptosis of cells in vitro, whether this phenomenon exists in vivo is still uncertain. Therefore, future studies are required to be performed in in vivo conditions. Besides, the specific mechanism was not studied. Future research would discuss whether caspase-3 and caspase-8 are involved in the autophagy process.
Conclusion
The results of this study show that the overexpression of Beclin1 in Hep-2 cells significantly inhibited cell proliferation and increased the rate of apoptosis in vitro. Beclin1 might be used as a target of gene therapy for LC.
Disclosure
The authors report no conflicts of interest in this work.
References
Salvador-Coloma C, Cohen E. Multidisciplinary Care of laryngeal cancer. J Oncol Pract. 2016;12:717–724. | ||
Devita VT, Lawrence TS, Rosenberg SA, De Pinho RA. DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2008. | ||
Jones AS, Fish B, Fenton JE, Husband DJ. The treatment of early laryngeal cancers (T1-T2 N0): surgery or irradiation? Head Neck. 2004;26(2):127–135. | ||
Tamura Y, Tanaka S, Asato R, et al. Therapeutic outcomes of laryngeal cancer at Kyoto University Hospital for 10 years. Acta Otolaryngol Suppl. 2007;557:62–65. | ||
Lozy F, Karantza V. Autophagy and cancer cell metabolism. Semin Cell Dev Biol. 2012;23(4):395–401. | ||
Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7(12):961–967. | ||
Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. | ||
Wang MC, Wu AG, Huang YZ, et al. Autophagic regulation of cell growth by altered expression of Beclin 1 in triple-negative breast cancer. Int J Clin Exp Med. 2014;8(5):7049–7058. | ||
Sun Y, Liu JH, Sui YX, et al. Beclin1 overexpression inhibits proliferation, invasion and migration of CaSki cervical cancer cells. Asian Pac J Cancer Prev. 2011;12(5):1269–1273. | ||
Wang W, Fan H, Zhou Y, Duan P, Zhao G, Wu G. Knockdown of autophagy-related gene BECLIN1 promotes cell growth and inhibits apoptosis in the A549 human lung cancer cell line. Mol Med Rep. 2013;7(5):1501–1505. | ||
Li X, Yan J, Wang L, et al. Beclin1 inhibition promotes autophagy and decreases gemcitabine-induced apoptosis in Miapaca2 pancreatic cancer cells. Cancer Cell Int. 2013;13(1):26. | ||
Weng J, Wang C, Wang Y, et al. Beclin1 inhibits proliferation, migration and invasion in tongue squamous cell carcinoma cell lines. Oral Oncol. 2014;50(10):983–990. | ||
Huang L, Wang S, Li SS, Yang XM. Prognostic significance of Beclin-1 expression in laryngeal squamous cell carcinoma. Pathol Oncol Res. 2013;19(4):771–777. | ||
Won KY, Kim GY, Kim YW, Song JY, Lim SJ. Clinicopathologic correlation of beclin-1 and bcl-2 expression in human breast cancer. Hum Pathol. 2010;41(1):107–112. | ||
Edinger AL, Thompson CB. Defective autophagy leads to cancer. Cancer Cell. 2003;4(6):422–424. | ||
Sun Y, Liu JH, Sui YX, et al. Beclin1 overexpression inhibits proliferation, invasion and migration of CaSki cervical cancer cells. Asian Pac J Cancer Prev. 2011;12(5):1269–1273. | ||
Lock R, Roy S, Kenific CM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22(2):165–178. | ||
Wang MC, Wu AG, Huang YZ, et al. Autophagic regulation of cell growth by altered expression of Beclin 1 in triple-negative breast cancer. Int J Clin Exp Med. 2015;8(5):7049–7058. | ||
Wu S, Sun C, Tian D, et al. Expression and clinical significances of Beclin1. LC3 and mTOR in colorectal cancer. Int J Clin Exp Pathol. 2015;8(4):3882–3891. | ||
Rohatgi RA, Janusis J, Leonard D, et al. Beclin1 regulates growth factor receptor signaling in breast cancer. Oncogene. 2015;34(42):5352–5363. | ||
Salminen A, Kaarniranta K, Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: impact on the aging process. Ageing Res Rev. 2013;12(2):520–534. | ||
Siddiqui MA, Mukherjee S, Manivannan P, Malathi K. RNase L cleavage products promote switch from autophagy to apoptosis by caspase-mediated cleavage of Beclin-1. Int J Mol Sci. 2015;16(8):17611–17636. | ||
Liu K, Shi Y, Guo X, et al. CHOP mediates ASPP2-induced autophagic apoptosis in hepatoma cells by releasing Beclin-1 from Bcl-2 and inducing nuclear translocation of Bcl-2. Cell Death Dis. 2014;5:e1323. | ||
Wirawan E, Vande WL, Kersse K, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. | ||
Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6(7):891–900. | ||
Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. Autophagy and apoptosis: where do they meet? Apoptosis. 2014;19(4):555–566. | ||
Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. | ||
Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. | ||
Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28(21):2128–2141. | ||
Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. | ||
Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29(12):1717–1719. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.