Back to Journals » Cancer Management and Research » Volume 11
OSacc: Gene Expression-Based Survival Analysis Web Tool For Adrenocortical Carcinoma
Authors Xie L , Wang Q, Nan F, Ge L, Dang Y , Sun X, Li N, Dong H, Han Y, Zhang G, Zhu W, Guo X
Received 14 May 2019
Accepted for publication 2 October 2019
Published 24 October 2019 Volume 2019:11 Pages 9145—9152
DOI https://doi.org/10.2147/CMAR.S215586
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Longxiang Xie,1 Qiang Wang,1 Fangmei Nan,1 Linna Ge,1 Yifang Dang,1 Xiaoxiao Sun,1 Ning Li,1 Huan Dong,1 Yali Han,1 Guosen Zhang,1 Wan Zhu,2 Xiangqian Guo1
1Bioinformatics Department of Predictive Medicine, Institute of Biomedical Informatics, Cell Signal Transduction Laboratory, Bioinformatics Center, Henan Provincial Engineering Center for Tumor Molecular Medicine, School of Software, School of Basic Medical Sciences, Henan University, Kaifeng 475004, People’s Republic of China; 2Department of Anesthesia, Stanford University, Stanford, CA, USA
Correspondence: Xiangqian Guo
Bioinformatics Department of Predictive Medicine, Institute of Biomedical Informatics, Cell Signal Transduction Laboratory, Bioinformatics Center, Henan Provincial Engineering Center for Tumor Molecular Medicine, School of Software, School of Basic Medical Sciences, Henan University, Kaifeng 475004, People’s Republic of China
Tel +86-371-23880585
Email [email protected]
Abstract: Gene expression profiling data with long-term clinical follow-up information are great resources to screen, develop, evaluate and validate prognostic biomarkers in translational cancer research. However, an easy-to-use interactive online tool is needed to analyze these profiling and clinical data. In the current work, we developed OSacc (Online consensus Survival analysis of ACC), a web tool that provides rapid and user-friendly survival analysis based on seven independent transcriptomic profiles with long-term clinical follow-up information of 259 ACC patients gathered from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. OSacc allows researchers and clinicians to evaluate the prognostic value of genes of interest by Kaplan–Meier (KM) survival plot with hazard ratio (HR) and log-rank test in ACC. OSacc is freely available at http://bioinfo.henu.edu.cn/ACC/ACCList.jsp.
Keywords: ACC, prognostic marker, over survival, web tool, KM plot
Introduction
Adrenocortical carcinoma (ACC) is a type of rare and aggressive malignancy with a poor 5-year over survival rate which is below 40%.1 Due to the limited treatment options for patients with locally advanced ACC and high risk of relapse even after major operative resection, identifying new prognostic biomarkers and therapeutic targets to improve the therapies for ACC patients is very important.2–4 In recent decades, clinical bio-samples have been extensively analyzed by gene microarray or RNA-Seq technologies.5,6 Although numerous public transcriptomic datasets with clinical information are available, it is very difficult for researchers or clinicians without bioinformatics training to quickly assess the prognostic value of putative biomarkers. Because getting, processing, filtering, and building survival model of large gene expression datasets are time-consuming and need professional programming skills. In addition, the reported prognostic biomarkers such as GLUT17 and PTTG18 in ACC need to be independently validated in multi-cohorts of different populations before being translated into clinical use.
To build a user-friendly tool that contains ACC gene expression profiles with clinical follow-up data to perform online ACC prognosis analysis, we collected the transcriptomic profiles and clinical information of 259 ACC patients from seven independent cohorts derived from TCGA and GEO to establish OSacc. OSacc is a web-based portal providing survival analysis and risk assessment in ACC datasets using gene symbol as input. This tool offers biomedical researchers and clinicians lacking bioinformatics training to quickly identify putative prognostic biomarkers from gene expression data in ACC. This web tool is available through http://bioinfo.henu.edu.cn/ACC/ACCList.jsp.
Materials And Methods
Data Collection
Messenger RNA (mRNA) expression profiling data with patients’ survival information were collected from GEO and TCGA. To gather the data from GEO, the searching keywords “Adrenocortical carcinoma” or “ACC” and “survival” were used in GEO database. The obtained TCGA data are Level 3 RNAseq data with clinical information of ACC patients. The detailed information of above data from GEO and TCGA are described in Table 1. Only dataset with ≥20 ACC samples was included in this study.
 |
Table 1 Clinical Characteristics Of The ACC Patients Used In OSacc |
OSacc Web Tool
OSacc is developed as previously described with minor modifications.9–13 In brief, OSacc contains two main components: storage and data analysis. A Java implementation was used to construct OSacc. OSacc used SQL Server database to provide the storage and management of the gene expression profiles and clinical follow-up data for ACC, and applied the Browser/Server architecture network management system to manage database. R package “survival” was used to perform Cox regression analysis to calculate the hazard ratio, 95% confidence intervals and p-value. R packages “survminer” and “ggplot2” were applied to plot the Kaplan–Meier curves (Figure S1). The web-based interface is shown in Figure 1.
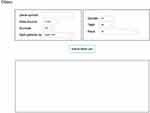 |
Figure 1 The web interface of OSacc. |
Searching Previous Known Prognostic Biomarkers In ACC
All known ACC prognostic biomarkers are searched in PubMed using the combined keywords “ACC” or “Adrenocortical carcinoma”, “prognostic” and “biomarkers”. Seven prognostic biomarkers were selected from 165 publications. The prognostic values of these previously reported prognostic biomarkers were determined in the combined datasets of ACC patients in OSacc.
Statistical Analysis
GraphPad Prism 8.0 (GraphPad Inc., La Jolla, CA, USA) software was used for statistical analysis. The mRNA expression of CENPF in ACC cancer tissues was compared with that in normal tissues, using a Wilcoxon rank-sum test (also called Mann–Whitney test) to calculate a p value. A value of p<0.05 was considered statistically significant.
Results
Clinical Characteristics Of ACC Datasets Used In OSacc
To obtain the microarray or RNAseq data of ACC with clinical survival information, we searched the GEO and TCGA database. Finally, a total of 259 ACC cases including 167 samples from 6 GEO datasets and 92 samples from TCGA datasets were collected. All of these samples are primary tumors. The median age of these patients is 48 years old and the ratio of male to female is 1:1.2. Two hundred and five patients have OS (overall survival), 34 patients have EFS (event-free survival), and 20 patients have PFS (progression-free survival). A summary of above ACC cohorts is shown in Table 1.
Application Of OSacc Web Tool
To measure the correlation between the gene of interest and survival rate, Kaplan–Meier plot was applied in OSacc. To use OSacc, users first input the query gene symbol, choose either one dataset or combined datasets, then select the median or other appropriate cutoff value of gene expression to categorize the ACC patients (Figure 2A). Combined datasets mean that each cohort was divided separately into strata by selecting the appropriate cutoff value, which are then pooled for survival analysis. Next, users select the specific survival terms including OS, EFS, PFS, DFI (disease-free interval), and PFI (progression-free interval) (Figure 2B). Users may be also interested in limiting their analysis in a subgroup of patients by selecting the optional confounding clinical factors such as TNM, gender, and race (Figure 2C–F). Finally, users click the “Kaplan–Meier plot” button, and the prognostic (Kaplan–Meier, KM) plots with HR, 95% CI and p value will be shown on the output web page. P value of all genes calculated by Univariate Cox analysis in OSacc are shown in Table S1.
 |
Figure 2 The screenshot of input parameter in OSacc (A–F). The options of main input parameters and clinical factors of OSacc. |
Validation Of Previously Published ACC Prognostic Biomarkers
To evaluate the performance of OSacc, the prognostic values of 7 ACC prognostic biomarkers were collected from PubMed (shown in Table 2), these 7 reported prognostic biomarkers include 4 unfavorable and 3 favorable ones. To test the OSacc functions, OS was selected as survival term and analyzed in the combined datasets. The cutoff value of splitting the patients was set as upper 25% vs lower 75%. The calculated HR (hazard ratio) and p value of these prognostic biomarkers are shown in Table 2. The analysis results in OSacc showed that the prognostic performances of 5 genes (i.e. GLUT1, PTTG1, MCT2, RARRES2 and Mki67) are in accordance with previous reports, while prognostic values of two genes (i.e. MCT1 and IGFBP2) are not the same as previously reported.
 |
Table 2 Validation Of The 7 Previous Reported Prognostic Biomarkers In OSacc |
Discovery Of Putative Prognostic Biomarkers In OSacc
In addition to validating the reported prognostic markers in independent clinical datasets, OSacc also can be employed to discover novel prognostic markers in ACC. According to the results of univariate Cox analysis in datasets of TCGA, GSE10927, GSE19750 and GSE33371, we selected the genes with p value consistently less than 0.05 in multiple datasets for following screening of potential prognostic biomarkers. CENPF (Centromere protein F), a key protein associated with the centromere-kinetochore complex, satisfies this criterion. CENPF plays a crucial role in chromosomal segregation during mitosis, and its overexpression has been previously reported in several types of solid tumors. Dai et al found that CENPF is highly expressed in HCC (Hepatocellular carcinoma) compared to control tissues, and its overexpression is significantly related with serum AFP, advanced differentiation stage and a shorter OS.14 In addition, functional experiments demonstrated that downregulation of CENPF inhibits the cell proliferation, formation of colonies and induces tumor growth in nude mice, suggesting its critical role in driving HCC tumorigenesis.14 In 2018, Shahid et al demonstrated that CENPF can regulate PC (Prostate cancer) metabolism by modulating pyruvate kinase M2 phosphorylation signaling.15 A recent study showed that downregulation of CENPF can change the global metabolic profiles of PC cells and inhibit cell proliferation, indicating that CENPF may be a vital regulator of PC metabolism.16 Data-mining of gene expression profile from 4 GEO datasets (GSE10927, GSE12368, GSE90713 and GSE33371) using non-parametric statistical method showed that CENPF expression is significantly up-regulated in ACC tissues compared with normal control tissues (Figure S2A–D), suggesting that CENPF may be linked to pathological process in ACC.
Using OSacc, we firstly found that patients with high expressing CENPF had worse OS, while the low expression of CENPF group had better OS in TCGA (HR: 10.498, 95% CI: 4.509–24.440, p<0.0001), GSE10927 (HR: 6.990, 95% CI: 2.031–24.060, p=0.0020), GSE19750 (HR: 3.738, 95% CI: 1.047–14.349, p=0.042), and GSE33371 datasets (HR: 5.075, 95% CI: 1.568–16.420, p=0.007) (Figure 3A–D). Not surprisingly, in the combined datasets of TCGA, GSE10927, GSE19750 and GSE33371, high CENPF-expressing group also showed poor OS (HR: 5.636, 95% CI: 3.414–9.303, p<0.0001) (Figure 3E), suggesting that CENPF is a novel unfavorable prognostic biomarker for OS in ACC. The KM plot cannot be shown in the other three datasets, because GSE49280 does not have the probe for CENPF, GSE76019 and GSE76021 do not have OS items.
Discussion
In this study, we developed a prognosis analysis web tool OSacc that comprises published transcriptomic datasets with clinical information for ACC, and provides survival analysis of ACC patients based on gene expression. The performance of OSacc has been evaluated by seven reported prognostic biomarkers, and the test results showed that 71% (5 of 7) of these reported prognostic biomarkers were confirmed to be prognostic significant in OSacc. The reasons for the insignificance of the 2 genes in OSacc may be due to the use of different detection level/method (ELISA in literature) and data from different ethnic groups.
Because of the importance of prognostic biomarker development in cancers, a couple of prognostic web tools have been developed, such as Proggene,17 OncoLnc18 and cBioportral.19 Prognene is good in performing an extensive survey of prognosis in general cancer types, however it has limited cases of ACCs. OncoLnc was developed to assess the prognostic significance of non-coding genes but not the coding gene. More importantly, OSacc has integrated seven ACC cohorts and incorporated the clinical covariates including TNM, gender and race to provide more informative survival plots to the researchers. The different features between OSacc and cBioportal are: (1) OSacc can help users to limit the analysis in a subgroup of patients by selecting the factors including age, gender and TNM, but cBioportal cannot do this; (2) OSacc can analyze the prognostic significance of interesting gene using different cutoffs including median, quartile and trichotomy of the expression level, while cBioportal just has median cutoff; (3) cBioportal just used the single dataset TCGA-ACC for genes’ survival analysis while OSacc integrated TCGA and GEO datasets for prognosis analysis. However, there are limitations for OSacc. For example, although combing seven datasets into one pooled analysis can enlarge the number of ACC patients, the quite large differences in overall survival across the studies or heterogeneity in tissue source or different analysis platform might be questioned in the interpretation of prognosis analysis results.
To our knowledge, OSacc is the largest ACC dataset for ACC prognosis analysis. Although the current sample size is only 268 ACC samples available, we will keep adding more ACC data into OSacc database and update the functionality when new ACC expression profiling dataset is available.
Conclusion
OSacc is a free, publicly accessible web tool allowing biomedical researchers and clinicians lacking bioinformatics training to easily analyze the prognostic potency of the genes of interest in ACC.
Acknowledgments
This study is supported by Innovative National Natural Science Foundation of China (No.81602362), supporting grants of Henan University (no. 2015YBZR048; no. B2015151), Yellow River Scholar Program (no. H2016012), and Program for Innovative Talents of Science and Technology in Henan Province (no. 18HASTIT048), Projects for College Students in Henan University (no. 201819002), China Postdoctoral Science Foundation (no. 2017M62237) and Henan Postdoctoral Foundation (no. 001702052), Program for Science and Technology Development in Henan Province (no.162102310391, no.172102210187), Program for Scientific and Technological Research of Henan Education Department (no.14B520022), Program for Young Key Teacher of Henan Province (2016GGJS-214), Kaifeng Science and Technology Major Project (18ZD008), supporting grant of Bioinformatics Center of Henan University (no. 2018YLJC01). Longxiang Xie and Qiang Wang are co-first authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Allolio B, Fassnacht M. Adrenocortical carcinoma: clinical update. Int J Clin Endocrinol Metab. 2006;91:2027–2037. doi:10.1210/jc.2005-2639
2. Libé R. Adrenocortical carcinoma (ACC): diagnosis, prognosis, and treatment. Front Cell Dev Biol. 2015;3:45. doi:10.3389/fcell.2015.00045
3. Xie L, Dang Y, Guo J, et al. High KRT8 expression independently predicts poor prognosis for lung adenocarcinoma patients. Genes. 2019;10:36. doi:10.3390/genes10010036
4. Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845. doi:10.1038/nrc1739
5. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE. 2013;8:e74250. doi:10.1371/journal.pone.0074250
6. Wang F, Yan Z, Lv J, et al. Gene expression profiling reveals distinct molecular subtypes of esophageal squamous cell carcinoma in Asian populations. Neoplasia. 2019;21:571–581. doi:10.1016/j.neo.2019.03.013
7. Fenske W, Voelker H-U, Adam P, et al. Glucose transporter GLUT1 expression is an stage-independent predictor of clinical outcome in adrenocortical carcinoma. Endocr Relat Cancer. 2009;16:919. doi:10.1677/ERC-08-0211
8. Demeure MJ, Coan KE, Grant CS, et al. PTTG1 overexpression in adrenocortical cancer is associated with poor survival and represents a potential therapeutic target. Surgery. 2013;154:1405–1416. doi:10.1016/j.surg.2013.06.058
9. Wang Q, Xie L, Dang Y, et al. OSlms: a web server to evaluate the prognostic value of genes in leiomyosarcoma. Front Oncol. 2019;9.
10. Xie L, Wang Q, Dang Y, et al. OSkirc: a web tool for identifying prognostic biomarkers in kidney renal clear cell carcinoma. Future Oncol. 2019. doi:10.2217/fon-2019-0296
11. Zhang G, Wang Q, Yang M, et al. OSblca: a web server for investigating prognostic biomarkers of bladder cancer patients. Front Oncol. 2019;9. doi:10.3389/fonc.2019.00466
12. Wang Q, Wang F, Lv J, et al. Interactive online consensus survival tool for esophageal squamous cell carcinoma prognosis analysis. Oncol Lett. 2019;18:1199–1206. doi:10.3892/ol.2019.10440
13. Wang Q, Zhang L, Yan Z, et al. OScc: an online survival analysis web server to evaluate the prognostic value of biomarkers in cervical cancer. Future Oncol. 2019. doi:10.2217/fon-2019-0412
14. Dai Y, Liu L, Zeng T, et al. Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma. Biochem Biophys Res Commun. 2013;436:711–718. doi:10.1016/j.bbrc.2013.06.021
15. Shahid M, Lee MY, Piplani H, et al. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metabolism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle. 2018;17:2802–2818. doi:10.1080/15384101.2018.1557496
16. Shahid M, Kim M, Lee MY, et al. Downregulation of CENPF remodels prostate cancer cells and alters cellular metabolism. Proteomics. 2019;1900038. doi:10.1002/pmic.v19.11
17. Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. doi:10.1186/1471-2407-14-970
18. Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67. doi:10.7717/peerj-cs.67
19. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi:10.1126/scisignal.2004088
20. Giordano TJ, Kuick R, Else T, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15:668–676. doi:10.1158/1078-0432.CCR-08-1067
21. Heaton JH, Wood MA, Kim AC, et al. Progression to adrenocortical tumorigenesis in mice and humans through insulin-like growth factor 2 and β-catenin. Am J Pathol. 2012;181:1017–1033. doi:10.1016/j.ajpath.2012.05.026
22. Assié G, Letouzé E, Fassnacht M, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607. doi:10.1038/ng.2895
23. Pinto EM, Rodriguez-Galindo C, Choi JK, et al. Prognostic significance of major histocompatibility complex class II expression in pediatric adrenocortical tumors: a St. Jude and Children’s Oncology Group Study. Clin Cancer Res. 2016;22:6247–6255. doi:10.1158/1078-0432.CCR-15-2738
24. Zheng S, Verhaak RGW, Giordano TJ, et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;29:723–736. doi:10.1016/j.ccell.2016.04.002
25. Pinheiro C, Granja S, Longatto-Filho A, et al. Metabolic reprogramming: a new relevant pathway in adult adrenocortical tumors. Oncotarget. 2015;6:44403. doi:10.18632/oncotarget.5623
26. Liu-Chittenden Y, Patel D, Gaskins K, et al. Serum RARRES2 is a prognostic marker in patients with adrenocortical carcinoma. Int J Clin Endocrinol Metab. 2016;101:3345–3352. doi:10.1210/jc.2016-1781
27. Patel D, Ellis R, Howard B, et al. Analysis of IGF and IGFBP as prognostic serum biomarkers for adrenocortical carcinoma. Ann Surg Oncol. 2014;21:3541–3547. doi:10.1245/s10434-014-3768-5
28. Kwok GT, Zhao JT, Glover AR, et al. Treatment and management of adrenal cancer in a specialized Australian endocrine surgical unit: approaches, outcomes and lessons learnt. ANZ J Surg. 2019;89:48–52. doi:10.1111/ans.15032
29. Duregon E, Molinaro L, Volante M, et al. Comparative diagnostic and prognostic performances of the hematoxylin-eosin and phospho-histone H3 mitotic count and Ki-67 index in adrenocortical carcinoma. Mod Pathol. 2014;27:1246. doi:10.1038/modpathol.2013.230
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

