Back to Journals » OncoTargets and Therapy » Volume 11
Oncogene mutational analysis in Chinese gastrointestinal stromal tumor patients
Authors Chen Q , Li R, Zhang Z, Deng Q, Li K, Wang H, Yang X, Wu Y
Received 26 October 2017
Accepted for publication 4 January 2018
Published 20 April 2018 Volume 2018:11 Pages 2279—2286
DOI https://doi.org/10.2147/OTT.S155214
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ingrid Espinoza
Video abstract presented by Qiong Chen.
Views: 265
Qiong Chen,1 Rong Li,2 Zhi-Gao Zhang,1 Qiao-Ting Deng,1 Kun Li,1 Hao Wang,1 Xue-Xi Yang,1 Ying-Song Wu1
1School of Laboratory Medicine and Biotechnology, Southern Medical University, Guangzhou, People’s Republic of China; 2Department of Tumor, Nanfang Hospital, Southern Medical University, Guangzhou, People’s Republic of China
Background: Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors and exhibit a high frequency of oncogenic KIT or PDGFRA mutations. Tyrosine kinase inhibitors (TKIs) have been mainly used in the treatment of GISTs bearing KIT/PDGFRA mutations. However, other mutation profiles have been found to affect the sensitivity to and effectiveness of TKIs in the treatment of GISTs.
Purpose: The aim of the present study was to describe the mutational status of multiple genes in GIST samples and to provide information for finding potential predictive markers of therapeutic targets in Chinese GIST patients.
Patients and methods: MassARRAY spectrometry was used to test 40 Chinese GIST patients for 238 mutations affecting 19 oncogenes.
Results: A total of 14 oncogenes with 43 mutations were detected in 38 samples, with a mutation frequency of 95%. Among these mutation samples, 26 GISTs were found for KIT or PDGFRA mutations, while 12 were KIT/PDGFRA wild-type. Approximately half of the GIST samples harbored multiple mutations. The most frequent mutations were found in KIT (62.5%), CDK4 (17.5%), NRAS (15%) and EGFR (12.5%). Other mutations included PIK3CA and AKT1 (10%), BRAF and ABL1 (7.5%), PDGFRA, ERBB2 and HRAS (5%), and AKT2, FLT3 and KRAS (2.5%). New mutated genes (CDK4, AKT2, FLT3, ERBB2, ABL1 and AKT1), a higher BRAF mutation frequency (7.5%) and new BRAF mutation sites (G464E) were found in Chinese GIST patients.
Conclusion: This study demonstrated useful mutations in a small fraction of Chinese GIST, but targeted therapeutics on these potential predictive markers need to be investigated in depth especially in Oriental populations.
Keywords: gastrointestinal stromal tumor (GIST), mutation, tyrosine kinase receptor, oncogene
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors originating in different parts of the digestive tract. GISTs have a characteristic morphology and biological continuum, and they are mostly incidentally discovered.1 Despite clinicopathological differences, most GISTs share a similar genetic profile, including oncogenic KIT or PDGFRA mutations.
Previous studies reported that KIT mutations are identified in 60%–85% of GISTs, while PDGFRA mutations are identified in 5%–10%.2 These mutations appear to be mutually exclusive, encoding a tyrosine kinase receptor type III.3,4 Thus, tyrosine kinase inhibitors (TKIs), such as imatinib, sunitinib, or sorafenib, are considered the main treatment for GISTs. However, previous reports suggest that KIT and PDGFRA mutations in GISTs mainly affect exons that code for functional domains of the KIT and PDGFRA receptors. Therefore, KIT and PDGFRA genotyping may be of value in predicting sensitivity to TKIs and selecting the optimal clinical treatment. For example, KIT exon 11 mutants respond well to imatinib, while exon 9 mutants (Ala502-Tyr503dup) are less sensitive to this TKI. PDGFRA exon 18 mutants (Asp842Val) are resistant to imatinib, and KIT exon 13 and 14 mutants are sensitive to sunitinib.5 However, KIT-negative GISTs present a true diagnostic challenge.
In addition, ~10%–15% of GISTs do not have detectable KIT or PDGFRA mutations (KIT/PDGFRA wild-type [WT] GISTs), suggesting that other molecular pathways may also be involved in the pathogenesis of these tumors. Mutations in NF6,7 and BRAF (V600E),8,9 or SDH complex genes,10 were detected in KIT/PDGFRA WT GISTs. Thus, GISTs are also characterized by five categories of oncogenic abnormalities, including KIT mutant, PGDFRA mutant, SDH-deficient, RAS/BRAF/NF1 mutant, or quadruple (KIT/PDGFRA/SDH/RAS-P) WT GISTs.11 The pathogenesis and underlying biology of quadruple WT GISTs is currently unknown. Further molecular and clinicopathological characterization of quadruple WT GISTs may help determine their prognosis as well as assist with the optimization of medical management, including clinical testing of novel therapies.11 Therefore, additional genetic testing may help identify therapeutic targets and develop novel therapeutic strategies for managing GISTs.
The aim of the present study was to describe the mutational status of multiple genes in GIST samples using the MassARRAY spectrometry platform. The results revealed 14 oncogenes with 43 mutations in 40 Chinese GIST patients, including 68.42% KIT or PDGFRA mutations and 31.58% KIT/PDGFRA WT GISTs. New mutation genes (CDK4, AKT2, FLT3, ERBB2, ABL1, and AKT1), a higher BRAF mutation frequency (7.5%), and new BRAF mutation sites (G464E) were identified in Chinese GIST patients. These mutation genes found in the present study may work as predictive markers for novel therapeutic targets in Chinese GIST patients.
Materials and methods
Patients and samples
Formalin-fixed paraffin-embedded samples from 40 patients with pathologically diagnosed GISTs were retrieved from the NanFang Hospital, Southern Medical University (Guangzhou, People’s Republic of China), between June 2006 and September 2011. All the cases were clinically treated with tumor resection. The clinical and follow-up data were updated in September 2011. This study was approved by the NanFang Hospital Ethics Committee, and written informed consent was obtained from all the participants.
Oncomutation detection
The OncoCarta panel (v1.0; Sequenom Inc., San Diego, CA, USA) was used to detect oncomutations in 40 GIST samples. This panel is a set of prevalidated assays for sensitive and efficient mutation screening by parallel analysis of 238 somatic mutations across 19 common oncogenes. The mutation types of each gene are listed in Table S1. DNA was extracted from each GIST sample using a QIAamp DNA formalin-fixed paraffin-embedded tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA (20 ng) was amplified using 24 sets of OncoCarta PCR primers. An extension reaction based on the OncoCarta extension primers was then performed. After salts were removed by the addition of a cation exchange resin, the reaction analyses were spotted onto a SpectroCHIP (Sequenom Inc.) and were analyzed using a MassARRAY matrix-assisted laser desorption/ionization time-of-flight mass spectrometry platform (Sequenom Inc.).
Analytical and statistical methods
Mutation data were analyzed by MassARRAY Typer Analyzer software 4.0.4.20 (Sequenom Inc.), using a cut-off mutation frequency of 1%. Automated mutation calls identified with the Typer software were generated using computational algorithms by quantifying the heights ratio of raw spectral peaks corresponding to the mutant and WT signals, noise-to-peak-height ratio, and area under the curve. In addition, the mutation report was manually reviewed by 3 investigators.
Results
Patient characteristics
Our study included 40 patients with GISTs who had undergone surgical resection. Mutation detection with the OncoCarta panel (ver.1.0; Sequenom Inc.) was performed in all the samples. The clinical characteristics of the patients are summarized in Table 1. The median age was 49 years (range, 20–84 years). Only 5% of these patients exhibited tumor recurrence or succumbed to the disease. A total of 80% of the patients were treated only with surgical resection and received no imatinib therapy, whereas 95% of the patients were insulin-like growth factor 1 receptor (IGF1R)-positive. All these results indicated that these tumors were low risk, with a low incidence of recurrence.
  | Table 1 Clinical characteristic of 40 GIST patients |
Mutation status in 40 GIST cases
Of the 40 GIST tumors, 38 (95%) were found to harbor oncogenic mutations. Of the 238 hotspot mutations in 19 common oncogenes, 14 oncogenes with 43 mutations were detected. The most frequent mutations were found in KIT (62.5%, 25/40), CDK4 (17.5%, 7/40), NRAS (15%, 6/40), and EGFR (12.5%, 5/40). Other mutations included PIK3CA and AKT1 (10%, 4/40), BRAF and ABL1 (7.5%, 3/40), PDGFRA, ERBB2, and HRAS (5%, 2/40), and AKT2, FLT3, and KRAS (2.5%, 1/40). The identified mutations are outlined in Figure 1.
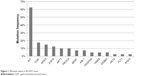  | Figure 1 Mutation status in 40 GIST cases. |
A total of 12 (30%) cases were found to be KIT/PDGFRA WT GISTs, including 4 cases with 2 or 3 coexisting mutations and 8 cases with a single mutation (Table 2). Sample 805823 harbored multiple mutations in ABL1 (E255K), AKT1 (rs11555435), and PIK3CA (E545K). Sample 707660 had two mutations in BRAF (G464E) and HRAS (G13S). Sample 8071414 harbored two mutations in ABL1 (T315I) and CDK4 (R24C). Sample 610972 harbored two mutations in ABL1 (G250E) and NRAS (G12D).
  | Table 2 Mutation analysis of GIST wild types |
The profiles of 26 cases with KIT or PDGFRA mutations are shown in Table 3. Also, most of the cases harbored multiple mutations.
  | Table 3 Mutation profiles of GIST in KIT or PDGFRA mutations |
Discussion
Cancer genetic information may provide important reference data for clinical diagnosis and treatment. Our research aimed to provide such information for identifying novel therapeutic targets by analyzing the mutational status of Chinese GIST patients for 238 hotspot mutations in 19 common oncogenes. A total of 43 mutations in 14 oncogenes were detected in 38 samples, with an overall mutation frequency of 95%. This result is consistent with a previous study reporting a single center’s experience with 275 GIST cases, among which mutations were identified in 93.8% of the cases.12 A total of 26 GISTs were detected for KIT or PDGFRA mutations, while 12 were found to be KIT/PDGFRA WT GISTs.
KIT is a cytokine receptor that belongs to the type III receptor tyrosine kinase family. It is structurally similar to PDGFRs, colony-stimulating factor-1 receptor, and fms-like tyrosine kinase. It has been reported that GISTs are generally positive for CD117 (c-kit) and are primarily caused by activating mutations in KIT or PDGFRA. Previous studies have demonstrated that KIT mutations are found in 60%–85% of GISTs, while PDGFRA mutations are found in 5%–10%.2 In the present study, the KIT and PDGFRA mutation frequencies were 62.5 and 5%, respectively, which is consistent with previous reports. The most common mutation of PDGFRA (D842V) was not identified in the present study; on the contrary, T674I (exon 14) and D1071N (exon 22) were identified. PDGFRA T674I is an imatinib-resistant type of PDGFRA, and this mutation status may provide useful information for the clinical treatment of GISTs.
KIT/PDGFRA WT GISTs are another type of GIST without KIT and PDGFRA mutations (10%–15%), in which the responsible pathogenetic pathways remain unknown. In our study, a high frequency (30%) of KIT/PDGFRA WT GISTs was detected among the 40 GIST samples, with 3 mutations in CDK4 and ABL1, 2 mutations in AKT1, PIK3CA, and NRAS, and 1 mutation in EGFR, HRAS, KRAS, ERBB2, and BRAF. Mutational analysis revealed that KRAS and ABL1 mutations were only detected in KIT/PDGFRA WT GISTs, and all ABL1 mutations were part of a multiple mutation status (results not shown).
It is well established that the RAS/RAF/ERK pathway plays an important role in tumor development, and KRAS, HRAS, and NRAS are the main components of the RAS/RAF/ERK pathway. Mutations in these genes occur in at least one-third of all human cancers, with KRAS mutations being the most common.13–15 In the present study of Chinese patients with GISTs, mutations of KRAS, NRAS, and HRAS were also detected. Among the 40 GISTs, 1 case (2.5%) of a KRAS G12C mutation was identified, which did not occur simultaneously with KIT, PDGFRA, or BRAF mutations. This mutation site differed from that reported by Hechtman et al16 in 2015, where one case with a KRAS G12V mutation was detected among 267 GISTs. Furthermore, 2 cases (5%) of HRAS mutations and 6 cases (15%) of NRAS mutations were detected among the 40 GISTs, whereas 1 HRAS mutation (G13S) and 2 NRAS mutations (G12D) were harbored by KIT/PDGFRA WT GISTs. It was previously reported that KRAS, NRAS, and HRAS mutations are scarce in GISTs.17 Although our results support that KRAS and HRAS mutation are scarce in GISTs, NRAS mutations were detected at a higher frequency among Chinese GIST patients. This result suggests that the role of NRAS mutations may differ among various populations, and it may play a key role in the RAS/RAF/ERK pathway in Chinese GIST patients.
EGFR mutation is one of the most important targets for biological therapy, particularly in non-small-cell lung cancer and colorectal cancer.18 However, there are very few literature on EGFR mutation in GISTs.19 In the present study, 4 EGFR mutations (D770_N771insG, T790M, and S752I/F) were detected among the 40 GISTs. Among these mutations, D770_N771insG and T790M occurred together with KIT, NRAS, or AKT1 mutations, whereas only the S752I/F mutation was harbored by KIT/PDGFRA WT GISTs. This result may overturn the hypothesis of Shi et al19 that EGFR mutations are mutually exclusive with KIT, PDGFRA, KRAS, or BRAF mutations in primary GISTs. In addition, the EGFR mutation frequency detected in our study is higher compared with previous reports. Therefore, we hypothesized that GISTs may be candidates for anti-EGFR-targeted therapy.
BRAF mutations are common in cancer and represent the most frequent genetic events in malignant melanoma. Multiple studies reported BRAF mutation V600E in KIT/PDGFR WT GISTs.8,20,21 In the present study, 4 cases of BRAF mutations (L597S and G464E) were detected, and G464E coexisted with the HRAS mutation G13S in KIT/PDGFR WT GISTs. This result infers a higher BRAF mutation frequency (7.5%) and indicates the presence of new mutation sites in GISTs.
The P13K 110 α subunit encoded by PIK3CA, a downstream effector in the KIT signaling pathway, has been identified in different types of cancer. In GISTs, PIK3CA mutations were also reported in a recent study.22 Similarly, in the present study, 4 cases (10%) were found to harbor PIK3CA mutations (H1047Y, E542K, and E545K). All the mutation sites identified in the present study have been reported in association with other tumors. A previous study based on immunohistochemistry suggested that activation of the mTOR signaling pathway is characteristic in PDGFRA mutant and WT GISTs, rather than KIT mutant GISTs.23 In the present study, 2 cases harbored E542K and E545K hotspot mutations of PIK3CA in KIT/PDGFRA WT GISTs. Thus, PIK3CA mutations may play a role in WT GIST pathogenesis.
Cyclin-dependent kinase 4, encoded by the CDK4 gene, is a member of the cyclin-dependent kinase family, is also referred to as cell division protein kinase 4, and is a catalytic subunit of the protein kinase complex that is important for cell cycle G1 phase progression. CDK4 mutations are associated with tumor cell growth. However, there has been no report of this gene’s mutations in GISTs to date. In the present study, 7 cases (17.5%) harbored CDK4 mutations at R24C (2 hotspots of R24C and R24H in CDK4 were detected). Nishida et al24 reported that genotyping and cell cycle analysis may be crucial for GIST risk stratification. Analyzing the GIST risk classification among these CDK4 mutation cases, it was observed that all these cases were high- or intermediate risk. This result was consistent with the study by Nishida et al.24 Taking the function of CDK4 into consideration, it was hypothesized that cyclin-dependent kinase inhibitors for tumor cell quiescence (associated with CDK4 mutations) may be a new therapeutic target in GISTs.
In addition, AKT2, FLT3, and ERBB2 mutations, concurrently with KIT mutations, were separately observed in 3 different cases. A total of 4 cases (10%) harbored AKT1 mutations and 3 cases were ABL1 mutation-positive among KIT/PDGFRA WT GISTs. To the best of our knowledge, there has been no report of these mutations in GISTs to date. Thus, AKT2, FLT3, and ERBB2 mutations are rarely present in GISTs. However, AKT2, FLT3, ERBB2, ABL1, and AKT1 were reported to be associated with GIST therapy.25–27 Therefore, the mutations observed in the present study may provide useful information for the clinical treatment of GISTs.
Conclusion
The present study using MassARRAY spectrometry screened 238 mutations affecting 19 oncogenes in 40 Chinese GIST patients. Fourteen oncogene mutations were detected in the samples, including KIT, CDK4, NRAS, EGFR, PIK3CA, AKT1, BRAF, ABL1, PDGFRA, ERBB2, HRAS, AKT2, FLT3, and KRAS. Approximately half of the GIST samples harbored multiple mutations. A higher frequency of KIT/PDGFRA WT GISTs was detected in the present study. In addition, CDK4, EGFR, PIK3CA, NRAS, KRAS, ERBB2, and AKT1 were single-point mutations detected in KIT/PDGFRA WT GISTs. New mutation genes (CDK4, AKT2, FLT3, ERBB2, ABL1, and AKT1) were also identified in Chinese GIST patients, along with a higher BRAF mutation frequency (7.5%) and new BRAF mutation sites (G464E). It is noteworthy that, although new mutations were detected in Chinese GIST patients, the sample size was insufficient to draw definitive conclusions. Therefore, further studies with larger samples that screen for mutations in full-length sequences are required to confirm our results.
Disclosure
The authors report no conflicts of interest in this work.
References
Miettinen M, Lasota J. Gastrointestinal stromal tumors – definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438(1):1–12. | ||
Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–4349. | ||
Rubin BP. Gastrointestinal stromal tumours: an update. Histopathology. 2006;48(1):83–96. | ||
Gasparotto D, Rossi S, Bearzi I, et al. Multiple primary sporadic gastrointestinal stromal tumors in the adult: an underestimated entity. Clin Cancer Res. 2008;14(18):5715–5721. | ||
Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53(3):245–266. | ||
Salvi PF, Lorenzon L, Caterino S, Antolino L, Antonelli MS, Balducci G. Gastrointestinal stromal tumors associated with neurofibromatosis 1: a single centre experience and systematic review of the literature including 252 cases. Int J Surg Oncol. 2013;2013:398570. | ||
Gasparotto D, Rossi S, Polano M, et al. Quadruple-negative GIST is a sentinel for unrecognized neurofibromatosis type 1 syndrome. Clin Cancer Res. 2017;23(1):273–282. | ||
Jasek K, Buzalkova V, Minarik G, et al. Detection of mutations in the BRAF gene in patients with KIT and PDGFRA wild-type gastrointestinal stromal tumors. Virchows Arch. 2017;470(1):29–36. | ||
Falchook GS, Trent JC, Heinrich MC, et al. BRAF mutant gastrointestinal stromal tumor: first report of regression with BRAF inhibitor dabrafenib (GSK2118436) and whole exomic sequencing for analysis of acquired resistance. Oncotarget. 2013;4(2):310–315. | ||
Wang JH, Lasota J, Miettinen M. Succinate Dehydrogenase Subunit B (SDHB) is expressed in neurofibromatosis 1-associated gastrointestinal stromal tumors (Gists): implications for the SDHB expression based classification of Gists. J Cancer. 2011;2:90–93. | ||
Pantaleo MA, Nannini M, Corless CL, Heinrich MC. Quadruple wild-type (WT) GIST: defining the subset of GIST that lacks abnormalities of KIT, PDGFRA, SDH, or RAS signaling pathways. Cancer Med. 2015;4(1):101–103. | ||
Wang M, Xu J, Zhao W, et al. Prognostic value of mutational characteristics in gastrointestinal stromal tumors: a single-center experience in 275 cases. Med Oncol. 2014;31(1):819. | ||
Ma BB, Lui VW, Poon FF, et al. Preclinical activity of gefitinib in non-keratinizing nasopharyngeal carcinoma cell lines and biomarkers of response. Invest New Drugs. 2010;28(3):326–333. | ||
Hui AB, Lo KW, Teo PM, To KF, Huang DP. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol. 2002;20(3):467–473. | ||
Kratz CP, Schubbert S, Bollag G, Niemeyer CM, Shannon KM, Zenker M. Germline mutations in components of the Ras signaling pathway in Noonan syndrome and related disorders. Cell Cycle. 2006;5(15):1607–1611. | ||
Hechtman JF, Zehir A, Mitchell T, et al. Novel oncogene and tumor suppressor mutations in KIT and PDGFRA wild type gastrointestinal stromal tumors revealed by next generation sequencing. Genes Chromosomes Cancer. 2015;54(3):177–184. | ||
Toda-Ishii M, Akaike K, Suehara Y, et al. Clinicopathological effects of protein phosphatase 2, regulatory subunit A, alpha mutations in gastrointestinal stromal tumors. Mod Pathol. 2016;29(11):1424–1432. | ||
Troiani T, Napolitano S, Della CC, et al. Therapeutic value of EGFR inhibition in CRC and NSCLC: 15 years of clinical evidence. ESMO Open. 2016;1(5):e88. | ||
Shi SS, Wu N, He Y, et al. EGFR gene mutation in gastrointestinal stromal tumors. Histopathology. 2017;71(4):553–561. | ||
Agaram NP, Wong GC, Guo T, et al. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosomes Cancer. 2008;47(10):853–859. | ||
Agaimy A, Terracciano LM, Dirnhofer S, et al. V600E BRAF mutations are alternative early molecular events in a subset of KIT/PDGFRA wild-type gastrointestinal stromal tumours. J Clin Pathol. 2009;62(7):613–616. | ||
Lasota J, Felisiak-Golabek A, Wasag B, et al. Frequency and clinicopathologic profile of PIK3CA mutant GISTs: molecular genetic study of 529 cases. Mod Pathol. 2016;29(3):275–282. | ||
Sapi Z, Fule T, Hajdu M, et al. The activated targets of mTOR signaling pathway are characteristic for PDGFRA mutant and wild-type rather than KIT mutant GISTs. Diagn Mol Pathol. 2011;20(1):22–33. | ||
Nishida T, Omori T, Nakayama S, et al. Prognostic importance of cell-cycle activity and genotype in gastrointestinal stromal tumors. J Clin Oncol. 2011;29(suppl 15):e20501. | ||
Wang Q, Liu F, Wang B, et al. Discovery of N-(3-((1-Isonicotinoylpiperidin-4-yl)oxy)-4-methylphenyl)-3-(trifluoromethyl)benzamide (CHMFL-KIT-110) as a selective, potent, and orally available type II c-KIT kinase inhibitor for gastrointestinal stromal tumors (GISTs). J Med Chem. 2016;59(8):3964–3979. | ||
Zook P, Pathak HB, Belinsky MG, et al. Combination of imatinib mesylate and AKT inhibitor provides synergistic effects in preclinical study of gastrointestinal stromal tumor. Clin Cancer Res. 2017;23(1):171–180. | ||
Rausch JL, Boichuk S, Ali AA, et al. Opposing roles of KIT and ABL1 in the therapeutic response of gastrointestinal stromal tumor (GIST) cells to imatinib mesylate. Oncotarget. 2017;8(3):4471–4483. |
Supplementary material
  | Table S1 Mutations detected with OncoCarta |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
