Back to Journals » Infection and Drug Resistance » Volume 11
Novel single-nucleotide variations associated with vancomycin resistance in vancomycin-intermediate Staphylococcus aureus
Authors Lin L , Chang S, Ge M , Liu T, Lu J
Received 4 August 2017
Accepted for publication 2 November 2017
Published 18 January 2018 Volume 2018:11 Pages 113—123
DOI https://doi.org/10.2147/IDR.S148335
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
Lee-Chung Lin,1 Shih-Cheng Chang,1,2 Mao-Cheng Ge,1 Tsui-Ping Liu,1 Jang-Jih Lu1–3
1Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou, Taoyuan, Taiwan; 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan; 3Department of Medicine, College of Medicine, Chang Gung University, Taoyuan, Taiwan
Abstract: Prolonged vancomycin usage may cause methicillin-resistant Staphylococcus aureus to become vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA). Mechanisms of vancomycin resistance of VISA and hVISA are still unclear. In this study, analyses of nucleotide sequence variations in 30 vancomycin-sensitive S. aureus (VSSA), 41 hVISA and 16 VISA isolates revealed 29 single-nucleotide variations in 12 genes (fmtC, graR, graS, htrA, mecA, pbp2, pbp4, srtA, tcaA, upps, vicK and vraR) that are related to cell wall synthesis or the two-component system. Six of these 29 single-nucleotide variations were novel and resulted in the following amino acid changes: Q692E in FmtC; T278I, P306L and I311T in HtrA; and I63V and K101E in Upps. Since P306L and I311T in HtrA and I63V in Upps were present in the majority (76.7%–86.7%) of VSSA isolates, these three amino acid variations may not be associated with vancomycin resistance. The other three amino acid variations (T278I in HtrA, K101E in Upps and Q692E in FmtC) were present in the majority (87.5%–93.8%) of hVISA and VISA isolates, but only in a small number (22.9%–25.7%) of VSSA isolates, suggesting that they are associated with vancomycin resistance.
Keywords: MRSA, VSSA, hVISA, VISA
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading cause of nosocomial infection.1 Vancomycin is usually used to treat MRSA infections, but high-level vancomycin-resistant S. aureus (VRSA) has emerged mainly due to the acquisition of the vanA operon from Enterococci.2 There are also VRSA isolates with lower levels of vancomycin resistance, including vancomycin-intermediate S. aureus (VISA) and heterogeneous VISA (hVISA).3 The causes for the different levels of vancomycin resistance in MRSA are not completely clear.4,5
Identification of VISA and hVISA is conventionally done by determination of population analysis profile and area under the curve ratio (PAP/AUC). However, PAP/AUC is very labor-intensive and not practical for clinical diagnosis. Transcriptome and proteome analyses of MRSA isolates have revealed a link between nucleotide sequence variations of several genes and vancomycin resistance.6,7 These genes include those of the two-component systems, such as vraRS, graRS or walRK,6,8,9 and those involved in cell wall synthesis, such as upps, fmtC and srtA.9–11 Single-nucleotide variations (SNVs) of genes related to antibiotic resistance have also been described.8,9,12,13 These SNVs may allow differentiation between vancomycin-sensitive S. aureus (VSSA) and VISA.
Previous studies on hVISA and VISA strains have revealed 60 genes that may be associated with vancomycin resistance.4,9,10,14–23 We hypothesized that some nucleotide sequence variations in these genes correlate with the difference in vancomycin susceptibility of VSSA and VISA isolates. To test this hypothesis, we investigated the prevalence of SNVs of these 60 genes and found 29 SNVs (in 12 genes) that were more common in hVISA and VISA than in VSSA isolates. Among them, three novel amino acid sequence variations including Q692E in FmtC, T287I in HtrA and K101E in Upps proteins were found to be significantly associated with vancomycin resistance.
Materials and methods
Bacteria strains and growth conditions
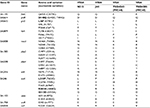
In total, 87 MRSA isolates were used in this study, including 72 from Chang Gung Memorial Hospital, 3 from National Taiwan University Hospital, 8 from Tri-Service General hospital (TSGH) and 2 each from Chi Mei Medical Center and China Medical University Hospital. These isolates were collected from 2009 to 2014. Detailed information of these isolates is presented in Table 1. All isolates were stored at −80°C and grown on tryptic soy agar plates at 37°C for 16 hours for the studies.
Whole-genome sequencing and polymerase chain reaction (PCR)
The whole genome of two VISA isolates (TSGH205 and TSGH243) was sequenced. Genomic DNA of each isolate was isolated and sonicated to generate fragments of 300–500 bp for construction of a DNA library, which was then subjected to Illumina next-generation sequencing. Raw sequence data generated were filtered and assembled for analysis using the CLC Genomics Workbench (QIAGEN, Venlo, the Netherlands). Genes selected for investigation of vancomycin resistance and PCR primers used to amplify these genes are shown in Table 2. PCRs were performed in a buffer containing 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2 and 50 mM KCl under the following conditions: 5 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C for denaturation, 30 seconds at 55°C for primer annealing and 1 minute at 72°C for extension, and 5 minutes at 72°C for final extension. PCR products were verified by sequencing.
  | Table 2 Primers used in this study |
Minimum inhibition concentration (MIC) E-tests
E-test was performed to determine the vancomycin MIC of VSSA, hVISA and VISA isolates. Cells of an overnight culture of each isolate were suspended in 0.9% NaCl solution, adjusted to 0.5 McFarland unit and spread evenly on a Mueller–Hinton agar plate. An E-test strip (bioMérieux, Durham, NC, USA) was placed on the agar plate, which was then incubated at 35°C overnight to determine MIC values.
Population analysis profile/area under the curve
PAP/AUC was performed to identify hVISA and VISA isolates as previously described.24,25 Briefly, serial dilutions of the overnight culture of an isolate were plated on brain heart infusion agar plates containing different concentrations of vancomycin (0, 0.5, 1, 1.5, 2, 3, 4, 6 and 8 μg/mL). After incubation at 35°C for 48 hours, colony-forming units (CFUs) of the isolate were determined and graphed as log10CFU/mL value versus vancomycin concentration to calculate the AUC. Reference strains Mu3 and Mu50 were analyzed in an identical manner to serve as hVISA and VISA controls, respectively. To distinguish VSSA, hVISA and VISA, the ratio (PAP/AUC value) of the AUC of an isolate to the AUC of Mu3 was calculated. The following ratios were used for identification of VSSA, hVISA and VISA populations: VSSA, <0.9; hVISA, 0.9–1.3; VISA, >1.3.
Results
Nucleotide sequence variations in genes related to vancomycin resistance
The 87 MRSA isolates used in this study included 30 VSSA, 41 hVISA and 16 VISA isolates (Table 1). To test the hypothesis that some SNVs in the previously identified 60 genes (Table S1) correlate with the difference in vancomycin susceptibility of VSSA and VISA isolates, the nucleotide sequences of these 60 genes in these MRSA isolates were examined. To simplify the bioinformatics work, this examination was performed in a stepwise manner to gradually narrow down the scope of analysis. Genes with SNVs were first identified, followed by determination of the prevalence of the SNVs in the MRSA isolates, identification of novel SNVs and then correlation of these novel SNVs with vancomycin resistance.
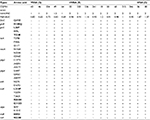
For the first step of analysis, the sequences of the 60 genes in two each of VSSA (N315 and JH1) and VISA (TSGH205 and TSGH243) isolates were aligned and compared. These four strains were selected because their entire genome had been sequenced. The two VSSA strains (N315 and JH1) were found to differ in nucleotide sequences in 13 genes, whereas the two VISA strains (TSGH205 and TSGH243) differed in nucleotide sequences in 25 genes, indicating that genes in the VISA isolates are more variable. Most of these sequence variations are SNVs. After excluding SNVs that were common in the two VSSA and two VISA strains, 29 SNVs in the following 12 genes were found in both VISA strains (TSGH205 and TSGH243): fmtC, graR, graS, htrA, mecA, pbp2, pbp4, srtA, tcaA, upps, vraR and vicK (Table 3). These genes have been shown to be related to the two-component system (graR, graS, vicK and vraR), cell membrane synthesis (tcaA), cell wall synthesis (mecA, pbp2, pbp4 and srtA) or vancomycin resistance (htrA, fmtC and upps).10,11
Prevalence of the 29 SNVs in VSSA, hVISA and VISA isolates
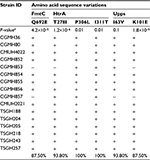
In the second step of analysis, the association of these 29 SNVs with vancomycin resistance was determined. To achieve the goal, the nucleotide sequences of the aforementioned 12 genes of 16 additional clinical isolates were compared, including five VSSA, nine hVIS and two VISA strains. Results showed that two VSSA, five hVISA and all VISA isolates had the same 29 SNVs. Six of these SNVs were novel and resulted in the following amino acid changes: Q692E in FmtC; T278I, P306L and I311T in HtrA; and I63V and K101E in Upps (Table 4).
Amino acid sequence variations in HtrA, FmtC and Upps associated with vancomycin resistance
In the third step of analysis, the role of these six novel SNVs in vancomycin resistance was then investigated by detecting their presence in 69 additional MRSA isolates. These isolates included 25 VSSA, 32 hVISA and 12 VISA isolates. Among the six amino acid variations, P306L and I311T in HtrA and I63V in Upps were present in the majority of VSSA isolates (P306L, 86.7%; I311T, 86.7%; I63V, 76.7%), as shown in Table 5, suggesting that these three amino acid variations are not associated with vancomycin resistance. The other three amino acid variations (T278I in HtrA, K101E in Upps and Q692E in FmtC) were found to be present in the majority of hVISA and VISA isolates, but only in a small number of VSSA isolates. The percentages of isolates with these sequence variations were the following: HtrA T278I: 87.8% hVISA, 93.8% VISA and 25.7% VSSA isolates; Upps K101E: 87.8% hVISA, 87.5% VISA and 22.9% VSSA isolates; and FmtC Q692E: 87.8% hVISA, 87.5% VISA and 25.7% VSSA isolates (Tables 5–7).
Discussion
Previous studies have revealed 60 genes that may be related to vancomycin resistance,6,9 suggesting that vancomycin resistance is a complex phenomenon and is mediated not by a single gene but by a group of genes. Among these 60 genes, 41 were found to be associated with vancomycin resistance by expression analysis;10,14,16,20–23 8 were found by overexpression, deletion or insertion mutations;10,15,18,19 11 were found by analysis of SNVs.4,9,11,17,20 In this study, we found 29 SNVs in the following 12 genes: fmtC, graR, graS, htrA, mecA, pbp2, pbp4, srtA, tcaA, upps, vraR and vicK (Table 3). Six of these 29 SNVs in fmtC, htrA and upps are novel. Since we only examined the sequences of these 12 genes in a limited number (87) of MRSA isolates, it is conceivable that other isolates may have other important sequence variations that were not detected in this study.
Of the 12 genes with SNVs, graR, graS, vicK and vraR are related to the two-component system.9 The others (mecA, pbp2, pbp4, srtA, tcaA, fmtC, htrA and upps) are more closely associated with vancomycin resistance. The mecA gene encodes the penicillin-binding protein 2a (PBP2a), which plays a role in resistance to beta-lactam antibiotics. Previous studies have shown that vancomycin treatment triggers deletions in the mecA gene.26,27 The N146K, N204K and G246E mutations in the MecA protein that we found in this study are located near or in the dimerization domain of PBP2a (KEGG database; http://www.kegg.jp/ssdb-bin/ssdb_motif?kid=sav:SAV0041) and would cause conformational changes of PBP2a. A recent study showed that these mutations (N146K, N204K and G246E) are highly associated with ceftaroline resistance,28 suggesting that they are also related to vancomycin susceptibility.
The pbp2 and pbp4 genes encode other penicillin-binding proteins (PBPs), which are also major components of the cell wall. The N-terminal domain of these PBPs encodes a glycosyltransferase, which catalyzes glycan chain polymerization from lipid II. Their C-terminal domain encodes a transpeptidase, which cross-links glycan chains.29 In addition to the glycosyltransferase and transpeptidase domains, PBP4 also has a domain encoding a carboxypeptidase, which hydrolyzes the C-terminal d-Ala-D-Ala peptide bond of the precursor of peptidoglycan.30 Previous studies have shown that vancomycin treatment increases the expression of pbp2, but decreases the expression of pbp4 in S. aureus.31 It has been shown that pbp4 mutations result in a decrease in its carboxypeptidase activity, leading to increased production of D-Ala-D-Ala termini in peptidoglycan and decreased vancomycin-binding affinity.31 The sequence variation S189T in PBP4, we found in this study, is located in its carboxypeptidase domain (KEGG database; http://www.kegg.jp/ssdb-bin/ssdb_motif?kid=sav:SAV0642) and conceivably would reduce its activity.
Previous studies on PBP2 have found that mutations in the transpeptidase domain of PBP2 increase the susceptibility of S. aureus to ceftizoxime.32 The C197Y, A420V and A557T mutations in PBP2 found in this study are located in its transpeptidase and transglycosylase domains (KEGG database; http://www.kegg.jp/ssdb-bin/ssdb_motif?kid=sav:SAV1450). The C197Y mutation has been shown to be associated with the susceptibility of S. aureus to ceftaroline,33 which is effective for treatment of hVISA or VISA infections.34 Although the other two mutations have not been investigated, it is conceivable that they will alter the activity of PBP2.
The srtA gene encodes sortase A, which is involved in the modification of cell surface proteins.35 Transcriptome analysis revealed a decreased expression of srtA by vancomycin treatment.10 The N167K mutation identified in this study is located in the sortase domain (KEGG database; http://www.kegg.jp/ssdb-bin/ssdb_motif?kid=sav:SAV2528) and, thus, would decrease its activity.
The tcaA gene encodes a membrane protein that has been shown to be associated with glycopeptide resistance.36 Structural analysis showed that amino acid residues 195–322 of the TcaA protein contain an OprB porin domain (http://www.kegg.jp/ssdb-bin/ssdb_motif?kid =sauf:X998_2340) of the ABC transporter, which plays a major role in carbohydrate uptake. Previous studies have shown that the ABC transporter is also involved in bacterial multidrug resistance.37 Since the five amino acid variations (L218P, Y237H, T262S, R283H and G312D) in the TcaA protein we found are all located in its OprB porin domain, it is conceivable that these mutations will affect vancomycin resistance.
In this study, six novel SNVs were found in fmtC, htrA and upps genes that have not been shown to be associated with vancomycin resistance. Three of them (Q692E in FmtC, T287I in HtrA and K101E in Upps) were correlated with vancomycin resistance (Tables 5–7). The htrA gene encodes a serine protease of the DegP family.38 Previous studies have shown that HtrA can suppress the production and secretion of bacteriocin by Streptococcus pneumonia.39 Bacteriocin-producing enterococci and staphylococci also have been described.40–42 There have been no reports on the relationship between bacteriocin production and vancomycin resistance. Bacteriocin production has been shown to augment niche competition by enterococci in the gastrointestinal tract.43 It is unknown whether any of the MRSA isolates examined in this study produces bacteriocin. Therefore, the mechanisms by which HtrA mediates vancomycin resistance remain to be investigated. The T278I mutation is predicted to be located in the coil structure of HtrA (http://www.ebi.ac.uk/interpro/protein/Q5HH63). It is possible that this mutation alters the secondary structure and thus the enzymatic activity of HtrA, leading to vancomycin resistance.
The upps gene encodes undecaprenyl pyrophosphate synthase, which catalyzes the formation of undecaprenyl pyrophosphate (UPP) by condensing farnesyl pyrophosphate with eight molecules of isopentenyl pyrophosphate.44 UPP is a lipid carrier during peptidoglycan synthesis. Inhibition of Upps has been shown to suppress cell wall synthesis and decrease the susceptibility of bacteria to vancomycin.11 Structural prediction (EMBL-EBL; http://www.ebi.ac.uk/interpro/protein/A0A1D4QTF1) revealed that Upps has a dimer interface residue at amino acid position 101. Thus, the K101E variation discovered in this study very likely would destabilize Upps, leading to vancomycin resistance.
The fmtC (also called mprF) gene encodes phosphatidylglycerol lysyltransferase that mediates lysinylation of phosphatidylglycerol.45 Mutations in fmtC in S. aureus and Bacillus subtilis have been shown to cause a decrease in the production of lysyl-peptidoglycan, thus increasing negative charges of the cell membrane and reducing the susceptibility of bacteria to vancomycin and daptomycin.46,47 The Q692E mutation, that we found in this study, is located in the IPR02430 domain (EMBL-EBL; http://www.ebi.ac.uk/interpro/entry/IPR024320), which is involved in the transfer of the lysyl group from l-lysyl-tRNA to membrane-bound peptidoglycan. This mutation very likely will disrupt this process, resulting in vancomycin resistance.
Limitations of this study include limited number of MRSA isolates examined and lack of detailed information of infections caused by the isolates, correlation between gene expression levels and vancomycin resistance, and functional studies of the SNVs. Therefore, the possibility that the SNVs discovered in this study may not completely correlate with vancomycin resistance of MRSA isolates still exists.
Conclusion
We have identified 29 SNVs that are more prevalent in hVISA and VISA than in VSSA isolates. Through sequence comparison of 87 isolates, we demonstrated that three novel SNVs in htrA, upps and fmtC genes are associated with vancomycin resistance. Since other environmental factors such as coinfections and patient’s conditions may also affect vancomycin resistance, allelic replacement or complementation of these SNVs needs to be performed to confirm the roles of these mutations in vancomycin resistance.
Acknowledgments
We thank Dr Chao-Hung Lee for editing the manuscript. This work was supported by grants from Chang Gung Memorial Hospital (CMRPG3D1382 and CMRPG3F1721) and the Ministry of Science and Technology, Taiwan (MOST-104-2320-B-182A-005-MY3 and MOST 105-2811-B-182A-004).
Disclosure
The authors report no conflicts of interest in this work.
References
Deresinski S. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin Infect Dis. 2005;40(4):562–573. | ||
Weigel LM, Clewell DB, Gill SR, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302(5650):1569–1571. | ||
Limbago BM, Kallen AJ, Zhu W, Eggers P, McDougal LK, Albrecht VS. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J Clin Microbiol. 2014;52(3):998–1002. | ||
Chen CJ, Lin MH, Shu JC, Lu JJ. Reduced susceptibility to vancomycin in isogenic Staphylococcus aureus strains of sequence type 59: tracking evolution and identifying mutations by whole-genome sequencing. J Antimicrob Chemother. 2014;69(2):349–354. | ||
Wang WY, Lee SY, Chiueh TS, Lu JJ. Molecular and phenotypic characteristics of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus isolates from patients with septic arthritis. J Clin Microbiol. 2009;47(11):3617–3623. | ||
Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(11):5845–5851. | ||
Peleg AY, Miyakis S, Ward DV, et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7(1):e28316. | ||
Cui L, Neoh HM, Shoji M, Hiramatsu K. Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(3):1231–1234. | ||
Yoo JI, Kim JW, Kang GS, Kim HS, Yoo JS, Lee YS. Prevalence of amino acid changes in the yvqF, vraSR, graSR, and tcaRAB genes from vancomycin intermediate resistant Staphylococcus aureus. J Microbiol. 2013;51(2):160–165. | ||
McAleese F, Wu SW, Sieradzki K, et al. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J Bacteriol. 2006;188(3):1120–1133. | ||
Lee YH, Helmann JD. Reducing the level of undecaprenyl pyrophosphate synthase has complex effects on susceptibility to cell wall antibiotics. Antimicrob Agents Chemother. 2013;57(9):4267–4275. | ||
Gardete S, Kim C, Hartmann BM, et al. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 2012;8(2):e1002505. | ||
Howden BP, Stinear TP, Allen DL, Johnson PD, Ward PB, Davies JK. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52(10):3755–3762. | ||
Cui L, Lian JQ, Neoh HM, Reyes E, Hiramatsu K. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49(8):3404–3413. | ||
Seidl K, Stucki M, Ruegg M, et al. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob Agents Chemother. 2006;50(4):1183–1194. | ||
Jansen A, Turck M, Szekat C, Nagel M, Clever I, Bierbaum G. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol. 2007;297(4):205–215. | ||
Mwangi MM, Wu SW, Zhou Y, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104(22):9451–9456. | ||
Renzoni A, Kelley WL, Barras C, et al. Identification by genomic and genetic analysis of two new genes playing a key role in intermediate glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(3):903–911. | ||
Schulthess B, Meier S, Homerova D, et al. Functional characterization of the sigmaB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob Agents Chemother. 2009;53(5):1832–1839. | ||
Shoji M, Cui L, Iizuka R, et al. WalK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55(8):3870–3881. | ||
Zhao Y, Verma V, Belcheva A, Singh A, Fridman M, Golemi-Kotra D. Staphylococcus aureus methicillin-resistance factor fmtA is regulated by the global regulator SarA. PLoS One. 2012;7(8):e43998. | ||
Chen H, Liu Y, Zhao C, et al. Comparative proteomics-based identification of genes associated with glycopeptide resistance in clinically derived heterogeneous vancomycin-intermediate Staphylococcus aureus strains. PLoS One. 2013;8(6):e66880. | ||
Sun H, Yang Y, Xue T, Sun B. Modulation of cell wall synthesis and susceptibility to vancomycin by the two-component system AirSR in Staphylococcus aureus NCTC8325. BMC Microbiol. 2013;13:286. | ||
Lu JJ, Lee SY, Hwa SY, Yang AH. Septic arthritis caused by vancomycin-intermediate Staphylococcus aureus. J Clin Microbiol. 2005;43(8):4156–4158. | ||
Ho CM, Hsueh PR, Liu CY, et al. Prevalence and accessory gene regulator (agr) analysis of vancomycin-intermediate Staphylococcus aureus among methicillin-resistant isolates in Taiwan SMART program, 2003. Eur J Clin Microbiol Infect Dis. 2010;29(4):383–389. | ||
Adhikari RP, Scales GC, Kobayashi K, Smith JM, Berger-Bachi B, Cook GM. Vancomycin-induced deletion of the methicillin resistance gene mecA in Staphylococcus aureus. J Antimicrob Chemother. 2004;54(2):360–363. | ||
Noto MJ, Fox PM, Archer GL. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52(4):1221–1229. | ||
Schaumburg F, Peters G, Alabi A, Becker K, Idelevich EA. Missense mutations of PBP2a are associated with reduced susceptibility to ceftaroline and ceftobiprole in African MRSA. J Antimicrob Chemother. 2016;71(1):41–44. | ||
Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32(2):234–258. | ||
Lavollay M, Arthur M, Fourgeaud M, et al. The beta-lactam-sensitive D,D-carboxypeptidase activity of Pbp4 controls the L,D and D,D transpeptidation pathways in Corynebacterium jeikeium. Mol Microbiol. 2009;74(3):650–661. | ||
Boyle-Vavra S, Yin S, Challapalli M, Daum RS. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob Agents Chemother. 2003;47(3):1028–1036. | ||
Leski TA, Tomasz A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol. 2005;187(5):1815–1824. | ||
Fernandez R, Paz LI, Rosato RR, Rosato AE. Ceftaroline is active against heteroresistant methicillin-resistant Staphylococcus aureus clinical strains despite associated mutational mechanisms and intermediate levels of resistance. Antimicrob Agents Chemother. 2014;58(10):5736–5746. | ||
Werth BJ, Steed ME, Kaatz GW, Rybak MJ. Evaluation of ceftaroline activity against heteroresistant vancomycin-intermediate Staphylococcus aureus and vancomycin-intermediate methicillin-resistant S. aureus strains in an in vitro pharmacokinetic/pharmacodynamic model: exploring the “seesaw effect”. Antimicrob Agents Chemother. 2013;57(6):2664–2668. | ||
Becker S, Frankel MB, Schneewind O, Missiakas D. Release of protein A from the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2014;111(4):1574–1579. | ||
Maki H, McCallum N, Bischoff M, Wada A, Berger-Bachi B. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48(6):1953–1959. | ||
Wilson DN. The ABC of ribosome-related antibiotic resistance. MBio. 2016;7(3):e00598-16. | ||
Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J Bacteriol. 2004;186(16):5258–5266. | ||
Kochan TJ, Dawid S. The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol. 2013;195(7):1561–1572. | ||
del Campo R, Tenorio C, Jimenez-Diaz R, et al. Bacteriocin production in vancomycin-resistant and vancomycin-susceptible Enterococcus isolates of different origins. Antimicrob Agents Chemother. 2001;45(3):905–912. | ||
dos Santos Nascimento J, dos Santos KR, Gentilini E, Sordelli D, de Freire Bastos Mdo C. Phenotypic and genetic characterisation of bacteriocin-producing strains of Staphylococcus aureus involved in bovine mastitis. Vet Microbiol. 2002;85(2):133–144. | ||
Ceotto H, Nascimento Jdos S, Brito MA, Bastos Mdo C. Bacteriocin production by Staphylococcus aureus involved in bovine mastitis in Brazil. Res Microbiol. 2009;160(8):592–599. | ||
Kommineni S, Bretl DJ, Lam V, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526(7575):719–722. | ||
Oldfield E. Targeting isoprenoid biosynthesis for drug discovery: bench to bedside. Acc Chem Res. 2010;43(9):1216–1226. | ||
Peschel A, Jack RW, Otto M, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193(9):1067–1076. | ||
Mishra NN, Yang SJ, Sawa A, et al. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(6):2312–2318. | ||
Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother. 2011;55(9):4326–4337. |
Supplementary material
  | Table S1 Comparative analysis gene list |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.