Back to Journals » Cancer Management and Research » Volume 13
Normal Basal Epithelial Cells Stimulate the Migration and Invasion of Prostate Cancer Cell RM-1 by TGF-β1/STAT3 Axis in vitro
Authors Li CY , Chen CY , An JH , Wu JB, Shen H
Received 21 January 2021
Accepted for publication 1 April 2021
Published 7 May 2021 Volume 2021:13 Pages 3685—3697
DOI https://doi.org/10.2147/CMAR.S303122
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Bilikere Dwarakanath
Chun-Yan Li,1 Chun-Ya Chen,1 Jian-Hong An,1 Jian-Bin Wu,2 Hong Shen1,3
1South China University of Technology School of Medicine, Guangzhou Higher Education Mega Center, Guangzhou, 510006, Guangdong, People’s Republic of China; 2Department of Oncology, The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, 510407, People’s Republic of China; 3Department of Pathology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, Guangdong, 510515, People’s Republic of China
Correspondence: Hong Shen Tel +86 20-61648227
Email [email protected]
Aim: Basal epithelial cells are absent in distant prostate cancer. This study aimed to investigate whether basal epithelial cells could suppress migration and invasion of prostate cancer cells to become a new treatment strategy for prostate cancer.
Main Methods: Basal epithelial cells were identified by immunofluorescence with anti-p63. Wound healing assays or transwell assays were used to explore the effects of basal epithelial cells, TGF-β 1, SB431542 (inhibitor of TGF-β type I receptor) or stattic (inhibitor of phosphorylated STAT3) on migration or invasion of mouse prostate cancer cell (RM-1). Concentration of TGF-β 1 was measured by ELISA assay. HE staining was used to investigate cell morphology. Immunocytochemistry with anti-p63 was used to identify basal epithelial cells. Levels of STAT3, p-STAT3 (Ser727) and proteins associated with EMT were measured with Western blot assay. Cell proliferation was measured with MTT or CCK8 assay.
Results: Normal basal epithelial cells acquired from mouse prostate were specific to anti-p63 and more than 90%. Basal epithelial cells and RM-1 could both secrete TGF-β 1. Basal epithelial cells and TGF-β 1 promoted the migration and invasion of RM-1 through changing the cell morphology and up-regulating expression of ZEB1, N-cadherin, vimentin, snail and p-STAT3 (Ser727), at the same time down-regulating E-cadherin of RM-1. SB431542 strongly suppressed migration, invasion as well as the expressions of EMT relevant proteins and p-STAT3 (Ser727) of co-cultured RM-1. In addition, stattic suppressed proliferation, migration and invasion of non-treated RM-1 and co-cultured RM-1.
Conclusion: Our study suggests that normal basal epithelial cells might stimulate the migration and invasion of RM-1 by TGF-β 1/STAT3 axis which could be suppressed by inhibitor of TGF-β receptor and inhibitor of p-STAT3. So, basal epithelial cells might not become a treatment strategy for prostate cancer, but our results could provide some researching references for other diseases which include basal epithelial cells such as prostatic intraepithelial neoplasia, prostatic hyperplasia, cervical cancer, or urinary bladder cancer.
Keywords: normal basal epithelial cells, RM-1, migration, invasion, TGF-β 1, STAT3
Introduction
TGF-β isoforms include TGF-β1, TGF-β2 and TGF-β3, but TGF-β1 is the most abundant.1 TGF-β1, a suppressor in normal cells whereas a promoter in cancer cells,2 plays a role in epithelial mesenchymal transition (EMT), cells migration and invasion.3 Normal basal epithelial cells secrete active significant amounts of TGF-β1.4 Signal transducer and activator of transcription 3 (STAT3), a cytoplasmic transcription factor, regulates cell proliferation, differentiation, apoptosis, angiogenesis, cell survival, cell migration and tumorigenesis.5,6 It has been reported that STAT3 was related to poor clinical prognosis of many carcinomas, including acute myeloid leukemia (AML), and solid tumors of the bladder, prostate, breast, brain, cervix, colon, esophagus, head-and-neck, kidney, liver, lung, ovary or pancreas.7 STAT3 has two important phosphorylation sites: a tyrosine residue at amino acid position 705 (Tyr705) and a serine phosphorylation site at position 727 (Ser727)8 which could be effective without the phosphorylation of Tyr705.6,9 Yu et al. suggested that STAT3 is a promising new target for cancer treatment.10 TGF-β1 could up-regulate expression of p-STAT3 to participate in progressions of some cancers, which has been consistently revealed in recent years.11–14
Prostate tubules and acinus consist of three types of epithelial cells: secretory luminal cells, neuroendocrine cells and basal epithelial cells.15 Basal epithelial cells are a blood–prostate barrier between luminal cells and basement membrane16 but are absent in prostate carcinoma.17–19 Loss of basal epithelial cells is a characteristic of prostate carcinoma20,21 and one of the diagnostic criteria of prostate cancer,19 suggesting that some essential connections might exist between basal epithelial cells and prostate cancer cells. So, we put out a question regarding whether basal epithelial cells could suppress migration or invasion of prostate cancer cells when basal epithelial cells are artificially added into a cultured system of prostate cancer cells. Through extensive reading of the medical literature, we found that the effects of basal epithelial cells on prostate cancer cells are still limited. One previous study reported that extracellular matrix of basal epithelial cell inhibited the proliferation of prostate cancer cell LNCaP by secreting TGF-β1.22 But another research found conditional medium of basal epithelial cell promoted the migration of BPH-1 by laminin-5 through the PI3K pathway.23 In conclusion, only a few studies have focused on the effects and mechanisms of normal basal epithelial cells on prostate cancer cells. Besides, previous authors just studied this effect through conditional medium or extracellular matrix, which could not simulate the natural relationship of basal epithelial cells and cancer cells.
In this study, we investigate the effect and mechanism of mouse normal basal epithelial cells on migration and invasion of mouse prostate cancer cells RM-1 through a co-cultured system, wound healing assay, transwell assay, ELISA, Western blot, HE staining, CCK8 assay and MTT assay. In the results, we found that basal epithelial cell might stimulate the migration and invasion of RM-1 through TGF-β1/STAT3 axis. So, we suggest that basal epithelial cells might not become a new treatment strategy for prostate cancer but our results might provide new research ideas for some early prostate diseases in which basal epithelial cells still exist such as benign prostatic hyperplasia and prostatic intraepithelial neoplasia (PIN). Besides, our research ideas could also provide new research inspirations for other cancers which can break through the basal epithelial cell layer and basement membrane to transfer.
Materials and Methods
Materials
Mouse prostate cancer cells RM-1 and mouse primary normal prostate basal epithelial cells were obtained from iCell Bioscience Inc. in Shanghai. All experiments involving mouse were approved by the animal center in South China University of Technology School of Medicine and the ethic number is 2018051. Inhibitor of phosphorylation of STAT3 (stattic, purity >98%) and inhibitor of TGF-β type I receptor (SB431542, IC50=94nM) were purchased form MedChemExpress company (USA). Fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 and RPMI-1640 basal medium were purchased from Gibco Company (USA). Recombinant TGF-β1 was purchased from Peprotech Company (USA). Primary antibody: anti-p63, anti-ZEB1, anti-E-cadherin, anti-vimentin, were purchased from Proteintech Company (Wuhan, China). Anti-snail, anti-N-cadherin, anti-STAT3, anti-p-STAT3 were purchased from WanLei Company (Beijing, China).
Cell Culture
Basal epithelial cells were cultured in (DMEM)/F12 medium supplemented with 5% FBS and 1% penicillin-streptomycin. RM-1 was cultured in RPMI-1640 medium with 10% FBS and 1% penicillin-streptomycin. Co-culture treated cells were cultured in (DMEM)/F12 medium including 5% FBS and 1% penicillin-streptomycin. All cells were incubated in an incubator with humidified atmosphere of 5% CO2 at 37°C.
Immunofluorescence
Basal epithelial cells were cultured on glass coverslips for 24 h. Glass coverslips were fixed in 4% paraformaldehyde followed by penetration with triton x-100 and blocked with goat serum. Glass coverslips were incubated with anti-p63 at 4°C overnight and washed with phosphate buffered saline (PBS) three times and then incubated with CoraLite594-conjugated secondary antibody for 2 h at normal temperature. Cells were stained with DAPI for 5 min and then photographed under fluorescence microscopy.
Cell Co-Cultured System
In wound healing assays, 1.0 x 105 RM-1 and 1.0 x 105 basal epithelial cells were mixed and cultured in a 6-well plate until confluence. In Western blot assays, basal epithelial cells were cultured in 6-well transwell upper chamber of which pore size was 0.4 µm, and RM-1 was in lower chamber for 72 h. In transwell assays, basal epithelial cells were cultured in 24-well transwell lower chamber, and RM-1 was in upper chamber of which pore size was 8 µm for 20 h. The above co-cultured systems have been sketched in Figure 1.
 |
Figure 1 Simple map of cell co-cultured system. |
Wound Healing Assays in vitro
RM-1 and basal epithelial cells (1:1) were co-incubated in the same 6-well plate for experiment group while only RM-1 was cultured for control group. The mono-layer cells were scratched in the middle of well plate with 200 µl pipette tips and washed with PBS to remove floating cells. PBS was replaced by (DMEM)/F12 without FBS. Images were photographed under microscope at 0 h and 24 h, respectively. Photos were quantified with Image J and the width of 24 h subtracted the width of 0 h for the migrated width.
Preparation of Conditional Medium of Basal Epithelial Cells
Basal epithelial cells which fused to 80% were washed with PBS three times and cultured with 4 mL (DMEM)/F12 without FBS. After 24 h incubation, the conditional medium was collected and centrifuged at 2000 r/min for 5 min and then stored at −80°C.
Cell Migration Assay
Cell migration assay was performed in 24-well transwell chamber of which pore size was 8 µm. 1.0 x 105 RM-1 was cultured in upper compartment with 200 µl (DMEM)/F12 without FBS. Lower chambers had 500 µl (DMEM)/F12 added with 10% FBS. 1.0 x 105 basal epithelial cells were cultured in lower chamber of co-culture groups.24 Conditional medium group had 500 µl medium including 80% (DMEM)/F12, 10% FBS and 10% conditional medium added to lower chamber. After incubation for 20 h, non-migrated cells in the upper compartment were removed with cotton swabs. Cells which penetrated the membrane were fixed with methanol for 20 min and stained with 0.1% crystal violet for 15 min. Five random fields of per chamber were photographed and the cell numbers quantified for statistics analysis.
Cell Invasion Assay
Cell invasion assay was performed in 24-well transwell chamber of which pore size was 8 µm. Transwell upper chamber was made with a thin underlay on the base with 80 µl matrix gel and incubated at 37°C overnight. The next procedures performed were the same as the migration assay.
HE Staining of Cells Growing on Glass Coverslips
Sterile glass coverslips were put on the base of 6-well plates, and cells were cultured on the coverslips. After incubation at 37°C for 48 h, cells were fixed with 4% paraformaldehyde for 1 h and stained with hematoxylin for 20 s and then washed and blued in running water followed by staining with eosin for 3 min. Coverslips were dehydrated with graded alcohol, transparented with xylene and coverslipped with resin.
Immunocytochemistry with Anti-p63
Cells on the coverslips were fixed with 4% paraformaldehyde and penetrated with triton x-100 and then blocked with goat serum. Glass coverslips were incubated with anti-p63 (1:100) at 4°C overnight and washed with phosphate buffered saline (PBS) three times and then incubated with HRP-conjugated secondary antibody for 30 min. Cells were colored by DAB and then stained by hematoxylin.
MTT or CCK8
RM-1 (1000 cells per well) was cultured in 96-well plate with 100 µl growth medium. After incubation, all wells had 10 µl MTT added and incubated at 37°C for 4 h continually. Then, all medium was replaced by 100 µl dimethyl sulfoxide to dissolve the formazan and plates were shaken for 15 min. Optical absorbance was measured at 570 nm using an enzyme-linked immunosorbent assay reader. In CCK8 assays, 10 µl CCK8 was added per well and incubated at 37°C for 2 h and then measured at 450 nm. Results were expressed by the mean values of results from replicated wells.
Western Blot Assay
Cells were lysed with RIPA lysis buffer on the ice box. Protein concentrations were detected with BCA protein assay kit (Beyotime Biotechnology, Shanghai, China). Proteins were separated by electrophoresis on 10% SDS-polyacrylamide gel, transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Burlington, MA, USA), and blocked by 5% fat-free milk at room temperature for 2 h. The membranes were covered with primary antibodies and incubated at 4°C overnight (ZEB1, N-cadherin, E-cadherin, vimentin, snail, p-STAT3 (Ser727), STAT3, β-actin at 1:1000, 1:500, 1:2500, 1:2000, 1:1000, 1:1000, 1:300, 1:800 respectively). Then, membranes were incubated with HRP-labeled secondary antibody (1:20,000) at room temperature for 1 h after being washed with TBST three times. The proteins of interest were detected by ECL chemiluminescence. The data of blot bands were analyzed with Image-pro plus software.
Statistical Analysis
Data were expressed as mean±SD followed by analysis using SPSS 17.0 software. Student’s t test was applied to analyze the difference between two groups. A p value lower than 0.05 represents statisticalsignificance. (*), (**), (***) indicate p<0.05, p<0.01, p<0.001 respectively. Each assay was performed independently at least three times. All graphs were made using GraphPad Prism 5.0.
Results
Identification of Basal Epithelial Cells Acquired from Mouse Prostate
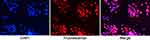
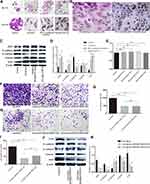
To identify the specificity and purity of mouse normal prostate basal epithelial cells, we performed cell immunofluorescence on the cell glass coverslips with anti-p63, and Figure 2 shows that the basal epithelial cells we isolated were specific to anti-p63 at more than 90%.
Mouse Normal Basal Epithelial Cells Could Stimulate the Migration of RM-1
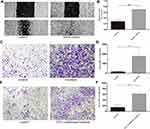
To investigate the effect of basal epithelial cells on migration of RM-1, we performed wound healing assay and migration assay. As shown in Figure 3A–D, basal epithelial cells significantly stimulated the migration of RM-1 in wound healing assay and migration assay. Ten percent of conditional medium of basal epithelial cells also promoted the migration of RM-1 (Figure 3E and F). In conclusion, it was demonstrated that normal basal epithelial cells could stimulate the migration of RM-1 directly or indirectly.
Mouse Normal Basal Epithelial Cells Could Promote the Invasion of RM-1
In order to explore the effect of basal epithelial cells on invasion of RM-1, we performed invasion assay with matrigel. Figure 4 reveals that basal epithelial cells and the conditional medium both stimulated the invasion of RM-1, which indicates that basal epithelial cells also promoted the invasion of RM-1 directly and indirectly.
Basal Epithelial Cell Could Secrete TGF-β1 and Additional TGF-β1 Stimulated the Migration and Invasion of RM-1 in vitro
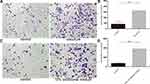
TGF-β1 is one of the most important factors in EMT. We performed TGF-β1 ELISA assay to measure the conditional medium of RM-1, basal epithelial cells and co-cultured system to explore whether basal epithelial cells could secrete TGF-β1. We performed migration and invasion assays to which were added 5 ng/mL TGF-β1 to investigate if TGF-β1 could promote the migration and invasion of RM-1. Figure 5A shows that normal basal epithelial cells and RM-1 could both secrete TGF-β1 and the concentration of co-culture group was higher. In the results shown in Figure 5B–D, addition of 5 ng/mL TGF-β1 stimulated the migration and invasion of RM-1.
TGF-β1 Was the Key Reason That Basal Epithelial Cells Promoted the Migration and Invasion of RM-1
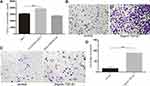
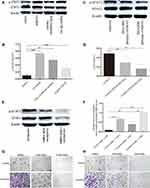
We investigated the effect of basal epithelial cells and TGF-β1 on EMT of RM-1 through the change of cell morphology as well as EMT relevant proteins of RM-1. Culturing RM-1 in (DMEM)/F12 with 5% FBS served as control group. Figure 6A shows that RM-1 grew centrally and cell morphologies were polygon or gravels in the control group, but RM-1 scattered relatively and the morphologies were spindle-shaped, needle-point or more elongated in co-cultured, 50% conditional medium treated and TGF-β1 (5 ng/mL) treated cells. Morphology of RM-1 also changed to mesenchymal-like in mixed-culture group (Figure 6B); the arrowhead indicates basal epithelial cells. In the results of Western blot (Figure 6C and D), some EMT relevant proteins such as ZEB1, N-cadherin, vimentin and snail expressed higher whereas E-cadherin expressed lower in co-cultured, 50% conditional medium treated and TGF-β1 (5 ng/mL) treated RM-1 than untreated RM-1. These results further illustrate that basal epithelial cells and TGF-β1 (5 ng/mL) could promote the migration and invasion of RM-1 in vitro.
To determine whether TGF-β1 is the key reason that basal epithelial cells promoted the migration and invasion of RM-1, we performed transwell assay and Western blot assay with or without SB431542 (2000 nM, 4000 nM), an inhibitor of TGF-β type I receptor (IC50=94nM). After incubation for 20 h with SB431542, we firstly performed CCK8 assays (Figure 6E) to study if different concentrations of SB432542 could kill RM-1 so that we could choose a suitable concentration to perform migration and invasion assay to decrease the effect of number of killer cells. As shown in Figure 6E, concentration of SB431542 from 500 nM to 4000 nM did not kill RM-1. Concentration of 2000 nM and 4000 nM strongly suppressed the migration (Figure 6F and G) and invasion (Figure 6H and I) of RM-1 which was co-cultured with basal epithelial cells. Besides, when SB431542 was added into co-cultured group, expressions of proteins associated with EMT (ZEB1, N-cadherin, vimentin, snail) were down-regulated as well as E-cadherin being up-regulated (Figure 6J and K). These results demonstrated that TGF-β1 secreted by basal epithelial cells was a key factor to promote the migration and invasion of RM-1.
TGF-β1 Secreted by Basal Epithelial Cells Might Promote Migration and Invasion of RM-1 Depending on the Phosphorylation of STAT3
To explore whether STAT3 is the downstream molecular basis of basal epithelial cells and TGF-β1 to promote the migration and invasion of RM-1, we performed Western blot to measure the expression levels of p-STAT3 of non-treated cells, co-cultured cells, 50% conditional medium treated cells, TGF-β1 treated cells, and co-cultured cells to which were added 2000 nM or 4000 nM SB431542. Figure 7A and B show that phosphorylation of STAT3 was up-regulated in co-cultured, 50% conditional medium treated and TGF-β1 treated cells but these treatments had no effect on the STAT3 protein levels. However, p-STAT3 protein was down-regulated in the co-cultured cells which were treated by SB431542, although it had no statistical significance in 2000 nM vs co-cultured group (Figure 7C and D). Taken together, these results illustrated that basal epithelial cells might promote migration and invasion of RM-1 through TGF-β1/STAT3 axis.
In order to investigate if p-STAT3 plays a role in the migration and invasion of untreated RM-1 and co-cultured RM-1, we performed Western blot assay to test whether stattic, an inhibitor of p-STAT3 (5 µM, 10 µM), could down-regulate the expression level of p-STAT3 and inhibit the migration and invasion of co-cultured RM-1. Before transwell assays, we firstly performed MTT assays to detect if stattic could kill RM-1 to influence the migration and invasion of RM-1. In the Figure 7F, stattic killed non-treated RM-1 and co-cultured RM-1. Besides, 5 µM and 10 µM stattic strongly suppressed the migration (Figure 7G) and invasion (Figure 7H) of non-treated RM-1 and co-cultured RM-1. Additionally, our result (Figure 7E) reveals stattic as an inhibitor of p-STAT3 could really down-regulate expression of p-STAT3 of co-cultured RM-1. Taken together, stattic suppressed the proliferation, migration and invasion of co-cultured RM-1 through inactivated p-STAT3, which reveals that phosphorylation of STAT3 is associated with proliferation, migration and invasion of RM-1.
Discussion
Main Backgrounds and Main Findings
Metastatic prostate cancer is one of the most frequent cancers among men and is an incurable disease, although the world is trying to find an effective treatment.25 Many authors have already worked on finding the mechanisms and reasons for metastasis of prostate cancer. Prostate epithelial cells mainly include luminal cells and basal epithelial cells. It is well known that human prostate cancer cells originate from luminal cells because prostate cancer cells display remarkable luminal phenotype and secrete luminal-derived prostate specific protein (PSA).26 Additionally, a previous study has indicated that focal absence of basal epithelial cells was relative to prostate cancer invasion.20 However, it has been reported that basal epithelial cells might be the origin of prostate cancer cells because basal epithelial cells respond to some pathways associated with disease progression such as PI3K signaling, FGF signaling and ERG1 signaling but luminal cells failed to respond.27 Thus, there are debates about the origin of prostate cancer cells and the function of basal epithelial cells.28 In this study, we researched the effect of basal epithelial cells on migration and invasion of RM-1 to investigate whether basal epithelial cells could suppress progression of prostate cancer to provide a new treatment strategy. However, we found that basal epithelial cells can stimulate migration and invasion of mouse prostate cancer cells (RM-1) directly or indirectly (Figures 3 and 4), which suggests that basal epithelial cells might not become a new therapy for prostate cancer.
TGF-β1 in Previous Studies and This Study
TGF-β1 which plays a role in EMT is secreted by many types of cells.29 Basal epithelial cells secrete active TGF-β1 but its growth is not influenced by this cytokine.4 Cancer-associated fibroblast,30 tumor-associated macrophages (TAMs)31 or other stromal cells32 secrete TGF-β1 to promote the migration or proliferation of human prostate cancer cells. In our study, the normal basal epithelial cells also secreted TGF-β1 and additional TGF-β1 promoted RM-1 to migrate and invade (Figure 5). In EMT, the morphology of cancer cells could change to mesenchymal phenotype.33 Besides, mesenchymal biomarkers such as vimentin and N-cadherin will up-regulate and epithelial cell marker E-cadherin will down-regulate,34 also the transcription factors associated to EMT like ZEB1/2, snail, twist, slug will up-regulate.35,36 Our results on effects of basal epithelial cells and TGF-β1 on morphology and EMT relevant proteins of RM-1 were consistent with previous studies (Figure 6A–D), which indicates that basal epithelial cells and TGF-β1 could promote the EMT of RM-1. Besides, literatures have shown that EMT is related to migration and invasion of cancer.34,37,38 So, our results can also reflect basal epithelial cells and TGF-β1 both stimulated the migration and invasion of RM-1 (Figure 6A–D). Next, we found that TGF-β receptor inhibitor (SB431542) strongly suppressed the migration (Figure 6F and G), invasion (Figure 6H and I) and some expressions of proteins associated with EMT as well as promoted the expression of E-cadherin (Figure 6J and K). Taken together, we concluded that basal epithelial cells could promote the migration and invasion through mainly secreting TGF-β1. However, 4000 nM SB431542 could inhibit the migration and invasion of co-cultured RM-1 incompletely and the inhibited rates of migrations and invasion are not significant statistically between 4000 nM SB431542 and 2000 nM SB431542 (Figure 6G and I). Thus, other molecular mechanism might exist besides TGF-β1.
STAT3 in Previous Studies and This Study
STAT3, which associates with poor prognosis of many types of cancers, is a common downstream effector of some cytokines such as IL-6, VEGF, and EGF.39 TGF-β1 up-regulates the expression of p-STAT3 to exert some biological functions, such as associating with prostate cancer resistance to enzalutamide,40 promoting EMT in human head and neck squamous cell carcinoma,12 prostate cancer,13 glioma cells14 and so on. In Figure 7, basal epithelial cells and TGF-β1 up-regulated the levels of p-STAT3 in RM-1 (Figure 7A and B) but SB431542 down-regulated the expression of p-STAT3 of co-cultured treated cells (Figure 7C and D). So, we could conclude that basal epithelial cells might promote the migration and invasion of RM-1 through the TGF-β1/STAT3 pathway. Besides, Figure 7G and H show that p-STAT3 inhibitor suppressed the migration and invasion of untreated RM-1 or co-cultured RM-1, suggesting that p-STAT3 played a role in the effect of basal epithelial cells on migration and invasion of RM-1. In previous studies, Yu et al. found that basal epithelial cells promoted the migration of BPH-1 in humans by the laminin 5/AKT pathway23 but Miniati et al. found that basal epithelial cells inhibited proliferation of human prostate cancer LNCaP through the TGF-β1 pathway.22 Thus, the interactions of basal epithelial cells with different prostate cancer cell lines might vary. The differences between these results and our results might due to the differing species and cell lines. In conclusion, we suggest that normal basal epithelial cells might promote the migration and invasion of RM-1 by TGF-β1/STAT3 axis, and the inhibitor of p-STAT3 or TGF-β type I receptor may offer new research ideas for earlier prostate disease in which basal epithelial cells still exist.
Findings and Limitations
Some findings of this study have not been reported in the past: Basal epithelial cells might promote the migration and invasion of RM-1 in vitro by TGF-β1/STAT3 axis and up-regulating proteins associated with EMT; Effect of basal epithelial cell on RM-1 was studied by co-cultured system; Stattic inhibited proliferation, migration and invasion of non-treated RM-1 and co-cultured RM-1. But this study also has some limitations. We have preliminarily studied and proved a TGF-β1/STAT3 axis in the effect of basal epithelial cells on migration and invasion of RM-1, but we have not explored it very deeply, especially the relationship between TGF-β1 and STAT3. Besides, human prostate cancer cell lines were divided into androgen-independent cell lines such as PC-3 and DU145 or androgen-dependent cell lines such as LNCaP.41 Although many biological functions and behavior of mouse cells are close to human cells and RM-1 is a better prostate cancer model,42 our results can not completely represent the effects of human basal epithelial cells on the human prostate cancer cells.
Research Significance
In a tumor micro-environment, the cell-cell interaction is complicated, because interactions exist between different molecules which are secreted by the cells. Basal epithelial cells and the basement membrane of prostate are the last line of defence from epithelial mesenchymal transition. Previous studies reported that basal epithelial cells still remain in benign prostatic hyperplasia43 and prostatic intraepithelial neoplasia (PIN).44 Progression of tumor is a process which is influenced by multiple factors. In our present study, we have found that basal epithelial cells might promote mouse prostate cancer cells to migrate and invade through the TGF-β1/STAT3 axis, suggesting that basal epithelial cells might not become a new treatment strategy for prostate cancer. But our results may offer new research ideas for earlier prostate disease in which basal epithelial cells are still present. Inhibiting this axis might be helpful to down-regulate the influence of basal epithelial cells on earlier prostate disease to slow down progression of disease. Besides, our research ideas could also provide new research references for some other cancers which can break through the basal epithelial cell layer and basement membrane to transfer, such as cervical cancer, urinary bladder cancer and so on.
Conclusion
In conclusion, our results show that basal epithelial cells might stimulate the migration and invasion of RM-1 through the TGF-β1/STAT3 axis which could be inhibited by TGF-β receptor inhibitor and the p-STAT3 inhibitor. We also found that TGF-β1 promoted the migration and invasion of RM-1 by up-regulating expression of p-STAT3 and proteins associated with EMT. We suggest that basal epithelial cells might not be a new therapeutic strategy for prostate cancer but the interaction of basal epithelial cells and RM-1 might provide a promising research idea for earlier prostate disease in which basal epithelial cells still exist. Besides, the research idea in our study could also provide some research inspirations for other cancers which can break through the basal epithelial cell layer and basement membrane to transfer.
Disclosure
All authors declare there are no conflicts of interest.
References
1. Chen W, Ten DP. Immunoregulation by members of the TGFbeta superfamily. Nat Rev Immunol. 2016;16(12):723–740. doi:10.1038/nri.2016.112
2. Massague J. TGFbeta in cancer. Cell. 2008;134(2):215–230. doi:10.1016/j.cell.2008.07.001
3. Khan MI, Hamid A, Adhami VM, Lall RK, Mukhtar H. Role of epithelial mesenchymal transition in prostate tumorigenesis. Curr Pharm Des. 2015;21(10):1240–1248. doi:10.2174/1381612821666141211120326
4. Salm SN, Koikawa Y, Ogilvie V, et al. Generation of active TGF-beta by prostatic cell cocultures using novel basal and luminal prostatic epithelial cell lines. J Cell Physiol. 2000;184(1):70–79. doi:10.1002/(SICI)1097-4652(200007)184:1<70::AID-JCP7>3.0.CO;2-U
5. Lee H, Jeong AJ, Ye SK. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019;52(7):415–423. doi:10.5483/BMBRep.2019.52.7.152
6. Qin HR, Kim HJ, Kim JY, et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68(19):7736–7741. doi:10.1158/0008-5472.CAN-08-1125
7. Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi:10.1038/nrclinonc.2018.8
8. You L, Wang Z, Li H, et al. The role of STAT3 in autophagy. Autophagy. 2015;11(5):729–739. doi:10.1080/15548627.2015.1017192
9. Galoczova M, Coates P, Vojtesek B. STAT3, stem cells, cancer stem cells and p63. Cell Mol Biol Lett. 2018;23(1):12. doi:10.1186/s11658-018-0078-0
10. Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi:10.1038/nrc3818
11. Wang B, Liu T, Wu JC, et al. STAT3 aggravates TGF-beta1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed Pharmacother. 2018;98:214–221. doi:10.1016/j.biopha.2017.12.035
12. Wang Y, Wu C, Zhang C, et al. TGF-beta-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett. 2018;436:52–62. doi:10.1016/j.canlet.2018.08.009
13. Cho KH, Jeong KJ, Shin SC, Kang J, Park CG, Lee HY. STAT3 mediates TGF-beta1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett. 2013;336(1):167–173. doi:10.1016/j.canlet.2013.04.024
14. Wang H, Tang F, Bian E, et al. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-beta. Mol Biol Rep. 2020;47(1):433–441. doi:10.1007/s11033-019-05146-2
15. Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967–2000. doi:10.1101/gad.1965810
16. El-Alfy M, Pelletier G, Hermo LS, Labrie F. Unique features of the basal cells of human prostate epithelium. Microsc Res Tech. 2000;51(5):436–446. doi:10.1002/1097-0029(20001201)51:5<436::AID-JEMT6>3.0.CO;2-T
17. Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568–571. doi:10.1126/science.1189992
18. Stoyanova T, Cooper AR, Drake JM, et al. Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc Natl Acad Sci U S A. 2013;110(50):20111–20116. doi:10.1073/pnas.1320565110
19. Magi-Galluzzi C. Prostate cancer: diagnostic criteria and role of immunohistochemistry. Mod Pathol. 2018;31(S1):S12–21. doi:10.1038/modpathol.2017.139
20. Man YG, Gardner WA. Focal degeneration of basal cells and the resultant auto-immunoreactions: a novel mechanism for prostate tumor progression and invasion. Med Hypotheses. 2008;70(2):387–408. doi:10.1016/j.mehy.2007.05.015
21. Diaz JI, Cazares LH, Corica A, John SO. Selective capture of prostatic basal cells and secretory epithelial cells for proteomic and genomic analysis. Urol Oncol. 2004;22(4):329–336. doi:10.1016/j.urolonc.2004.04.010
22. Miniati DN, Chang Y, Shu WP, Peehl DM, Liu BC. Role of prostatic basal cells in the regulation and suppression of human prostate cancer cells. Cancer Lett. 1996;104(2):137–144. doi:10.1016/0304-3835(96)04243-7
23. Yu HM, Frank DE, Zhang J, You X, Carter WG, Knudsen BS. Basal prostate epithelial cells stimulate the migration of prostate cancer cells. Mol Carcinog. 2004;41(2):85–97. doi:10.1002/mc.20041
24. Guo C, Huang T, Wang QH, et al. Monocarboxylate transporter 1 and monocarboxylate transporter 4 in cancer-endothelial co-culturing microenvironments promote proliferation, migration, and invasion of renal cancer cells. Cancer Cell Int. 2019;19:170. doi:10.1186/s12935-019-0889-8
25. Sur S, Steele R, Shi X, Ray RB. miRNA-29b inhibits prostate tumor growth and induces apoptosis by increasing bim expression. Cells-Basel. 2019;8(11).
26. Xin L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene. 2013;32(32):3655–3663. doi:10.1038/onc.2012.541
27. Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci U S A. 2010;107(6):2610–2615. doi:10.1073/pnas.0913873107
28. Lee SH, Shen MM. Cell types of origin for prostate cancer. Curr Opin Cell Biol. 2015;37:35–41. doi:10.1016/j.ceb.2015.10.002
29. Yang Y, Zhou X, Gao H, Ji SJ, Wang C. The expression of epidermal growth factor and transforming growth factor-beta1 in the stenotic tissue of congenital pelvi-ureteric junction obstruction in children. J Pediatr Surg. 2003;38(11):1656–1660. doi:10.1016/S0022-3468(03)00577-3
30. Sun DY, Wu JQ, He ZH, He MF, Sun HB. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-beta signaling pathway. Life Sci. 2019;235(2019):116791. doi:10.1016/j.lfs.2019.116791
31. Zhang D, Qiu X, Li J, Zheng S, Li L, Zhao H. TGF-beta secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle. 2018;17(24):2766–2778. doi:10.1080/15384101.2018.1556064
32. Costanza B, Umelo IA, Bellier J, Castronovo V, Turtoi A. Stromal modulators of TGF-beta in cancer. J Clin Med. 2017;6(1):7. doi:10.3390/jcm6010007
33. Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):e8. doi:10.1126/scisignal.2005189
34. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28(1–2):15–33. doi:10.1007/s10555-008-9169-0
35. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45.
36. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi:10.1016/j.jhep.2016.05.007
37. Odero-Marah V, Hawsawi O, Henderson V, Sweeney J. Epithelial-mesenchymal transition (EMT) and prostate cancer. Adv Exp Med Biol. 2018;1095:101–110.
38. Gheldof A, Berx G. Cadherins and epithelial-to-mesenchymal transition. Prog Mol Biol Transl Sci. 2013;116:317–336.
39. Ogata H, Chinen T, Yoshida T, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene. 2006;25(17):2520–2530. doi:10.1038/sj.onc.1209281
40. Liu Q, Tong D, Liu G, et al. Metformin reverses prostate cancer resistance to enzalutamide by targeting TGF-beta1/STAT3 axis-regulated EMT. Cell Death Dis. 2017;8(8):e3007. doi:10.1038/cddis.2017.417
41. Namekawa T, Ikeda K, Horie-Inoue K, Inoue S. Application of Prostate Cancer Models for Preclinical Study: advantages and limitations of cell lines, patient-derived xenografts, and three-dimensional culture of patient-derived cells. Cells. 2019;8(1).
42. Grant JF, Iwasawa T, Sinn HW, et al. Induction of protective immunity to RM-1 prostate cancer cells with ALVAC-IL-2/IL-12/TNF-alpha combination therapy. Int J Cancer. 2006;119(11):2632–2641. doi:10.1002/ijc.22220
43. Bonkhoff H, Stein U, Remberger K. The proliferative function of basal cells in the normal and hyperplastic human prostate. Prostate. 1994;24(3):114–118. doi:10.1002/pros.2990240303
44. Bostwick DG, Pacelli A, Lopez-Beltran A. Ultrastructure of prostatic intraepithelial neoplasia. Prostate. 1997;33(1):32–37. doi:10.1002/(SICI)1097-0045(19970915)33:1<32::AID-PROS6>3.0.CO;2-B
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.