Back to Journals » OncoTargets and Therapy » Volume 14
NGS-Based ctDNA Profiling After the Resistance of Second-Line Osimertinib for Patient with EGFR-Mutated Pulmonary Adenocarcinoma
Authors Li D, Yang D, Cui S, Pan E, Yang P, Dai Z
Received 10 May 2021
Accepted for publication 30 June 2021
Published 21 July 2021 Volume 2021:14 Pages 4261—4265
DOI https://doi.org/10.2147/OTT.S318250
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Dan Li,1,* DaFu Yang,1,* SaiQiong Cui,1 Evenki Pan,2 Peng Yang,2 ZhaoXia Dai1
1The Second Department of Thoracic Medical Oncology, The Second Hospital of Dalian Medical University, Dalian, Liaoning, People’s Republic of China; 2Nanjing Geneseeq Technology Inc., Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: ZhaoXia Dai
The Second Department of Thoracic Medical Oncology, The Second Hospital of Dalian Medical University, No. 467, Zhongshan Road, Shahekou District, Dalian, Liaoning, People’s Republic of China
Tel +8617709873617
Email [email protected]
Abstract: Osimertinib, a third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), is effective in EGFR T790M positive non-small-cell lung cancer (NSCLC). Despite the efficacy of osimertinib, patients inevitably develop resistance and the mechanisms of osimertinib resistance are heterogeneous. Here, we report that a lung adenocarcinoma patient with EGFR L858R mutation who was treated with second-line osimertinib therapy acquired multiple resistance to osimertinib by the non-invasive circulating tumor DNA (ctDNA) genotyping. This case provides the possible mechanisms of osimertinib resistance that occur during the disease progression and supports the longitudinal monitoring of ctDNA for the detection of novel acquired resistance and tumor heterogeneity.
Keywords: osimertinib, acquired resistance, EGFR mutation, lung adenocarcinoma, circulating tumor DNA
Introduction
In non-small lung cancer (NSCLC), epidermal growth factor receptor (EGFR) gene has been one of the earliest well-characterized oncogenic mutations, which occur in approximately 45% of Asian lung adenocarcinoma (ADC) patients, among which the most frequent are short in-frame deletions in exon 19 (Ex19del) and point mutations in exon 21 (L858R).1 EGFR tyrosine kinase inhibitor (TKI) monotherapies have been the standard treatment for lung cancers with activating EGFR mutations. More than one-half of patients with EGFR mutations progressing on first-generation EGFR-TKIs (gefitinib and erlotinib) and second-generation EGFR-TKI (afatinib) develop the EGFR T790M resistance mutation.2 Osimertinib is the third-generation EGFR-TKI that originally approved as second-line treatment for patients with acquired resistance to first- or second-generation EGFR-TKIs through the T790M secondary mutation. However, the Phase III FLAURA trial demonstrated that first-line osimertinib is superior to first-generation EGFR-TKIs in overall response rate, progression-free and overall survivals,3 leading to United States Food and Drug Administration (FDA) approval of osimertinib as the first-line EGFR-TKI in April 2018.
Although osimertinib has demonstrated high clinical efficacy, developing resistance is also inevitable. With the development of next-generation sequencing (NGS) technology, circulating tumor DNA (ctDNA) as a liquid biopsy has been applied to adapt systemic therapy and monitor patients’ tumor progression.4 Herein, we reported a lung ADC patient with EGFR mutation treated with osimertinib as second-line treatment and revealed multiple resistant mechanisms using NGS-based ctDNA profiling.
Case Report
A 77-year-old man was diagnosed with stage IV (T1N3M1) lung ADC with right middle lobe, right-sided pleural effusion and multiple bone metastasis in June 2016 (Figure 1A). EGFR L858R mutation was identified by amplification refractory mutation system-polymerase chain reaction (ARMS-PCR); thus, the patient was initially given icotinib (125mg, tid). After an initial partial response (PR) was achieved, the patient reached a progression-free survival (PFS) of 16 months. However, in October 2017, the left-side ureter was irregularly thickened and multiple nodules occurred in the retroperitoneum, which indicated progressive disease (PD). Then, he was switched to osimertinib, a third-generation EGFR-TKI, at a dose of 100mg qd. The pain of the lower back and left lower extremity significantly eased during the initial treatment of osimertinib. The patient achieved a stable disease (SD) of lung lesion, but a remarkable elevated level of serum carcinoembryonic antigen (CEA) was found (Figure 1B). Subsequently, three sequential plasma samples during the treatment course of osimertinib were subjected to NGS targeting of 139 cancer-relevant genes to monitor mutation profiles of ctDNA. The dynamic mutation profiles are presented in Table 1.
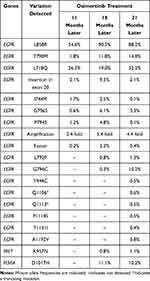 |
Table 1 Genetic Alterations Detected by ctDNA Sequencing During the Osimertinib Treatment Course |
In November 2018, the patient got progressed and the concomitant EGFR alterations including L858R (54.6%), T790M (1.8%), L718Q (26.3%), insertion in exon 20 (2.1%), I744M (1.7%), G796S (0.6%), P794S (1.2%), amplification (2.4 fold) and fusion (0.2%) was identified in the patients’ ctDNA from plasma. However, the patient refused chemotherapy and continued to take osimertinib. Seven months later, besides other EGFR alterations including L858R (90.5%), T790M (11.8%), L718Q (19.0%), insertion in exon 20 (9.5%), I744M (2.5%), G796S (6.1%), P794S (4.8%), amplification (5.4 fold) and fusion (2.2%), the NGS revealed some newly occurred mutations including EGFR L792P (0.8%), EGFR G796C (0.3%), MET K957N (0.8%) and PIK3CA D1017H (11.1%), in the plasma. In September 2019, to further monitor the resistant mutations, the NGS identified some mutations that had never occurred including Y944C (0.5%), truncated Q1106 (0.6%), truncated Q1113 (0.5%), P1114S (0.5%), T1131I (0.4%) and A1192V (0.8%). Unfortunately, the patient insisted on taking osimertinib and died in May 2020.
Discussion
Osimertinib, a third-generation EGFR TKI, selectively blocks the activated EGFR T790M mutation. Despite high efficacy of osimertinib, acquired resistance is a growing clinical challenge. The mechanisms of resistance to osimertinib have been demonstrated in previous reports. The most common EGFR-dependent and -independent mechanisms of resistance occur on C797S, MET-amplification and small cell lung carcinoma (SCLC) transformation.5,6 Most acquired resistant mutations to osimertinib we reported in this patient have also been reported by other groups, including C797S, L718Q, L792P, G796S and G796C.7,8 Mutation of C797S in EGFR, a cysteine to serine change at codon 797 located in the ATP-binding site, causes the loss of the covalent bond between osimertinib and the mutant EGFR.9 Both in vivo and vitro data showed that EGFR L718Q, the most common mutations in L718, affected the conformation of the EGFR-osimertinib complex.10 In silico modeling has demonstrated that a substitution in L718 residue within the ATP-binding site of the EGFR kinase domain can result in spatial restriction and hinder osimertinib-EGFR binding.11 Moreover, mutations in the L792 can sterically interfere with a methoxy group on the phenyl ring of osimertinib and disrupt its binding to EGFR kinase domain.12 Mutations in G796 have the potential to sterically interfere with the osimertinib–EGFR interaction, but different mutations in the G796 residue, such as G796R, G796C, G796S, G796D, may have different impact on the binding of osimertinib to EGFR.9 In addition, EGFR exon 20 insertion and EGFR gene amplification, which were found in our patient, has also been detected in some cases after failure of osimertinib treatment.13,14 Besides, we also found that I744M, P794S, Y944C, truncated Q1106, truncated Q1113, P1114S, T1131I, A1192V and EGFR fusion, but their roles in mediating resistance have not been established yet.
The activation of bypass pathways, aberrant downstream signaling or histologic transformation is ascribed as responsible for EGFR-independent mechanisms of resistance to osimertinib. MET gene amplification is the most frequent cause of bypass pathway activation.15 Rare mutations of MET in P97Q and I865F have also been reported.10 We identified MET K957N after osimertinib failure, but its significance and mechanism of resistance to osimertinib are still unknown. PIK3CA mutations can activate the PI3K pathway, in which E545K, E542K, R88Q, N345K and E418K occurred frequently and have been reported as acquired resistance to osimertinib.9 In our patient, PIK3CA D1017H was identified after progression on osimertinib.
We present comprehensive mutation profiles of an osimertinib-resistance lung cancer patient using cfDNA profiling. The development of targeting NGS has made it possible to identify gene alterations in ctDNA extracted from plasma samples,16 which can allow the investigation of a panel of multiple cancer‐related genes. Thus, liquid biopsy has emerged as a promising alternative to tissue biopsy for molecular monitoring of patients throughout disease. In our case, we demonstrated that longitudinal monitoring of ctDNA mutation profiles may be a powerful tool for the detection of tumor genetic evolution and EGFR‐TKI treatment in NSCLC patients. Further detailed studies are needed to validate the function of acquired multiple resistance to osimertinib.
Conclusion
We monitored the patient treated with osimertinib by serial ctDNA analysis to detect patients’ tumor progression and novel mutations, which provides possible multiple resistant mechanisms to anti-EGFR therapy and highlights the importance of ctDNA sequencing to explore novel resistance mechanisms with therapeutic implications, which may trigger the development of effective therapeutic strategies.
Data Sharing Statement
All datasets generated for this study are included in the manuscript.
Ethics Approval and Consent to Participate
The study was approved by the ethics committee of the Second Hospital of Dalian Medical University. The patient provided written informed consent to participate in this study. Written informed consent for the publication of this case report and accompanying images was obtained from the patient. We would like to thank the patient and his family for providing consent for publication. We also thank all staff involved in this case study.
Acknowledgments
We sincerely thank the patient for supporting our work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
Evenki Pan and Peng Yang are the employees of Nanjing Geneseeq Technology Inc., China. Other authors have no conflicts of interest to declare.
References
1. Shu Y, Wu X, Tong X, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7(1):583. doi:10.1038/s41598-017-00520-1
2. Nagano T, Tachihara M, Nishimura Y. Mechanism of resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and a potential treatment strategy. Cells. 2018;7(11):212. doi:10.3390/cells7110212
3. Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2019;382(1):41–50. doi:10.1056/NEJMoa1913662
4. Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020;1(3):276–290. doi:10.1038/s43018-020-0043-5
5. Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M–positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4(11):1527–1534. doi:10.1001/jamaoncol.2018.2969
6. Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol. 2018;29:i10–i19. doi:10.1093/annonc/mdx703
7. Lee J, Shim JH, Park WY, et al. Rare mechanism of acquired resistance to osimertinib in Korean patients with egfr-mutated non-small cell lung cancer. Cancer Res Treat. 2019;51(1):408–412. doi:10.4143/crt.2018.138
8. Zhang Q, Zhang XC, Yang JJ, et al. EGFR L792H and G796R: two novel mutations mediating resistance to the third-generation EGFR tyrosine kinase inhibitor osimertinib. J Thorac Oncol. 2018;13(9):1415–1421. doi:10.1016/j.jtho.2018.05.024
9. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121(9):725–737. doi:10.1038/s41416-019-0573-8
10. Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non–small cell lung cancer patients. Clin Cancer Res. 2018;24(13):3097–3107. doi:10.1158/1078-0432.CCR-17-2310
11. Bersanelli M, Minari R, Bordi P, et al. L718Q mutation as new mechanism of acquired resistance to AZD9291 in EGFR-mutated NSCLC. J Thorac Oncol. 2016;11(10):e121–123. doi:10.1016/j.jtho.2016.05.019
12. Ou S-HI, Cui J, Schrock AB, et al. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/G and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer. 2017;108:228–231. doi:10.1016/j.lungcan.2017.04.003
13. Papadimitrakopoulou V, Wu Y-L, Han J-Y, et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol. 2018;29:viii741. doi:10.1093/annonc/mdy424.064
14. Knebel FH, Bettoni F, Shimada AK, et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer. 2017;108:238–241. doi:10.1016/j.lungcan.2017.04.004
15. Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7(1):11815. doi:10.1038/ncomms11815
16. Kitazono S, Sakai K, Yanagitani N, et al. Barcode sequencing identifies resistant mechanisms to epidermal growth factor receptor inhibitors in circulating tumor DNA of lung cancer patients. Cancer Sci. 2019;110(10):3350–3357. doi:10.1111/cas.14153
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

