Back to Journals » International Journal of General Medicine » Volume 16
NEK6 Promotes the Progression of Osteosarcoma Through Activating STAT3 Signaling Pathway by Down-Regulation of miR-26a-5p
Authors Zhu M , Sun Y, Xue H, Wu G, Wang Z, Shi J, Ma J, Gu B, Yan X
Received 21 March 2023
Accepted for publication 19 June 2023
Published 4 July 2023 Volume 2023:16 Pages 2831—2848
DOI https://doi.org/10.2147/IJGM.S413461
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Min Zhu,1 Yuyu Sun,1 Huawei Xue,1 Gang Wu,1 Zhen Wang,1 Junfeng Shi,1 Jiye Ma,1 Baorong Gu,1 Xiaoling Yan2
1Department of Spine Surgery, Nantong Third People’s Hospital, Affiliated Nantong Hospital 3 of Nantong University, Nantong, People’s Republic of China; 2Department of Chemotherapy, Affiliated Hospital of Nantong University, Nantong, People’s Republic of China
Correspondence: Xiaoling Yan, Department of Chemotherapy, Affiliated Hospital of Nantong University, Nantong, 226001, People’s Republic of China, Email [email protected]
Background: Osteosarcoma is a malignant tumor originating from the skeletal system. There is no effective treatment other than surgery and chemotherapy, which seriously endangers the health of children and adolescents. NEK6 is a novel discovered Serine/Threonine protein kinase that can regulate cell cycle and activate several oncogenic pathways.
Methods: NEK6 expression in pan-cancer including sarcoma was evaluated using analysis tools of TIMER, UALCNA and GEPIA with TCGA database, and its association with overall survival in patients with sarcoma was also analyzed. TargetScan, tarbase, microT-CDS and Starbase online software were used to predict NEK6-targeted miRNAs, including miR-26a-5p. Tumor tissues from patients with osteosarcoma were collected for NEK6 and miRNA detection using RT-qPCR. NEK6 down-regulated by siRNAs or miR-26a-5p in osteosarcoma cells was detected by RT-qPCR, Western blot and Immunofluorescence staining assays. Effects of NEK6 knockdown on proliferation, migration, invasion and apoptosis of osteosarcoma cells were detected by CCK-8, wound healing, transwell and flow cytometry, respectively. The expressions of STAT3, metastasis and apoptosis-related genes were detected by Western blot.
Results: High expression of NEK6 and low expression of miR-26a-5p were lowly expressed in osteosarcoma and they were negative correlation. NEK6 has been confirmed as a direct target for miR-26a-5p. In addition, NEK6 down-regulated by siRNAs or miR-26a-5p led to inhibition of cell proliferation, migration and invasion while promoting cell apoptosis. The levels of phosphorylated STAT3 and metastasis genes (MMP-2, MMP-9) were inhibited, while apoptotic gene Bax was promoted and Bcl2 was inhibited by miR-26a-5p upregulation.
Conclusion: NEK6 can promote osteosarcoma progression via activating STAT3 signaling pathway, which is inhibited by miR-26a-5p, suggesting that NEK6 is a potential oncogene and miR-26a-5p is a suppressor of osteosarcoma. The strategy of inhibiting of NEK6 by miR-26a-5p may be an effective approach for osteosarcoma therapy.
Keywords: osteosarcoma, NEK6, miR-26a-5p, STAT3 signaling pathway
Introduction
Osteosarcoma is a malignant tumor of the primary skeletal system and developed from stromal cells, with a global incidence of 3.4 cases per million people per year.1 Osteosarcoma accounts for 56% of all bone tumors and is more common than Ewing’s sarcoma, chondrosarcoma, and chordoma.2 In 80% of patients, tumors occur in the long diaphyseal region, especially in the area of rapid bone growth. Approximately 40% of osteosarcomas originate in the femur, 20% in the tibia, and 10% in the humerus.3 Notably, patients over 25 years old exhibit a wide range of primary tumor sites, as 20% also exhibit axial bone and soft tissue malignancies.4
Currently, standard treatments for patients with osteosarcoma include surgical excision, radiation and chemotherapy. The current treatment regimen combines surgery with multiple chemotherapy modalities using a variety of cytotoxic agents, such as cisplatin, doxorubicin, high-dose methotrexate, and ifosfamide, both pre- and post-operatively.5 For recurrent diseases, surgical excision is superior to systemic therapy, while unresectable cases will be treated by systemic therapy or comprehensive therapy.6 In addition, radiotherapy can help eliminate small or minimal residual tumors when substantial surgical resection is not feasible.7 However, in most cases of osteosarcoma, radiotherapy is limited and its efficacy and indications are limited.8 Due to surgical excision, combined chemotherapy and targeted radiotherapy, the 5-year survival rate of patients without metastasis can be about 60% to 70%.9 However, metastasis and recurrence often result in a poor prognosis, so the 5-year survival rate for patients with metastatic or recurrent osteosarcoma is only 10% to 20%.10 Recent studies have shown that 10% to 15% of patients diagnosed with osteosarcoma will develop metastases.11 In patients with metastatic osteosarcoma, lung metastasis is the most common (about 74%), followed by bone metastasis (about 9%), and about 8% of patients have both bone and lung metastasis.12 Standards of treatment for osteosarcoma, most of which were established in the 1980s, include surgery, chemotherapy, and radiotherapy, and can achieve long-term survival in about 60% of patients with localized osteosarcoma.13 However, while the heterogeneity and underlying molecular mechanisms of osteosarcoma have been further studied as the biology of osteosarcoma is better understood, there have been no new breakthroughs in the treatment of osteosarcoma, especially in inoperable osteosarcoma patients who are insensitive or intolerant to chemotherapeutic agents, and what can be done is still limited. In addition, drug resistance to chemotherapy drugs is also a problem that hinders breakthroughs in the treatment of osteosarcoma.14 Therefore, clarifying the molecular mechanism of osteosarcoma and developing targeted drugs to improve the patients’ survival rate are the key issues to be solved urgently.
Never in mitosis gene A-related expressed kinase (NEK) family consists of 11 members that locate in different cellular regions, they are widely involved in cell biological processes such as cell cycle, cell aging, telomere maintenance and chemical resistance.15 NEK expression was lower in G1 phase of the cell cycle, increased in S phase and G2 phase, and decreased after the cells entered mitosis, and activity peaked in the S and G2 phases and assisted centrosome separation in the G2/M phase. Thus, in tumors, NEK overexpression leads to chromosomal instability and aneuploidy in cancer cells. Moreover, NEK overexpression also activates several oncogenic pathways, enabling normal cells to transform into cancer cells by acquiring proliferation, invasion and drug resistance.16 As one of the less studied kinase families, the exact molecular mechanism of NEK family in tumorigenesis remains unclear.17 NEK6 is an important member of the NEK family, and it is a novel discovered Serine/Threonine protein kinase that regulates cell cycle and plays an important role in regulating a variety of kinases and transcription factors in different signaling pathways.18 NEK6 that has been found to express in 133 breast cancer samples was higher than that in paracancer tissues, and the high expression of NEK6 was related to the histological grade, tumor size and TNM stage of breast cancer, indicating that NEK6 expression was an independent prognostic predictor of breast cancer, suggesting that it could be a potential therapeutic target for breast cancer.19 In addition, researchers have also found that NEK6 is highly expressed in prostate cancer, and overexpression of NEK6 stimulates cytoskeletal, differentiation, and immune signaling pathways.20 NEK6 is also highly expressed in hepatocellular carcinoma,21 colon cancer,22 and gastric cancer.23 However, the molecular mechanism of NEK6 in osteosarcoma has not been reported.
miRNAs are a class of small RNAs with a length of about 21 nucleotide (nt), and often play roles in mammalian cells as tumor suppressor or oncogenic miRNAs.24 Studies have reported that miR-26a-5p was low expressed in a variety of tumors and is closely related to the biological function of tumor cells, and is often used as a tumor suppressor miRNA.25–27
In this study, the roles and mechanism of NEK6 in osteosarcoma will be clarified, and the regulation of miR-26a-5p on NEK6, and the potential signaling pathway will be also explored. This study will provide experimental basis and theoretical research basis for the clinical diagnosis and treatment of osteosarcoma.
Materials and Methods
Bioinformatics Data Evaluation
Expression data from the Cancer Genome Atlas (TCGA) database were analyzed using Tumor IMmune Estimation Resource (TIMER),28 the University of ALabama at Birmingham CANcer (UALCNA)29 and Gene Expression Profiling Interactive Analysis (GEPIA),30 respectively. Patients with sarcomas were divided into two groups for Kaplan–Meier (KM) curve analysis of overall survival according to the median level of NEK6.
NEK6-targeted miRNAs were predicted using TargetScan (https://www.targetscan.org/vert_80/),31 tarbase (https://dianalab.e-ce.uth.gr/html/diana/web/index.php?r=tarbasev8),32 microT-CDS (https://dianalab.e-ce.uth.gr/tools),33 and Starbase (https://starbase.sysu.edu.cn/index.php).34 And the predicted miRNAs were further analyzed using Venn diagram online (http://www.ehbio.com/test/venn/#/).
Human Tissue Samples
Human osteosarcoma tissues were collected from 13 patients with osteosarcoma and normal para-carcinoma tissues were also collected between June 2019 and December 2020 at Affiliated Hospital of Nantong University (China). The collected tissues were rapidly frozen in liquid nitrogen and stored at −80 °C. The present study was approved by the Ethics Committee of Affiliated Hospital of Nantong University (China) and followed the guidelines described in the Helsinki Declaration. Written informed consent was obtained for tissue sample collection from each participant.
Cells Culture and Transfection
Human osteosarcoma cell lines (Saos-2, MG-63) and an osteoblast cell line (hFOB1.19) were purchased from National Collection of Authenticated Cell Cultures (Shanghai, China). Saos-2 cells were cultured in McCoy’s 5A Medium (Gibco) with 10% Fetal Bovine Serum (FBS) (Lonsera), and MG-63 cells were cultured in Minimum Essential Medium (MEM) (Gibco) with 10% FBS (Lonsera), hFOB1.19 cell was cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12) (Gibco) with 10% FBS (Lonsera) and 0.3 mg/mL Geneticin 418 (G418) (Gibco). In addition, human embryonic kidney 293T cells (ATCC; #CRL-1573) were cultured in DMEM containing 10% FBS (Lonsera). All cells were maintained at 37 °C with 5% CO2 in a humidified incubator.
The specific-targeted small interfering (si)RNAs were designed and used to knockdown NEK6 expression, and the siRNA that has no homology with human genes was used as negative control (si-NC), miR-26a-5p mimics were used to up-regulation endogenous of miR-26a-5p and scrambled sequence of miR-26a-5p mimic was used as negative control (mimics NC). The siRNAs and miRNA mimics were transfected into cells at the concentration of 20 μM using Lipofectamine® 3000 (Thermo Fisher Scientific) at 37 °C for 24 h according to manufacturer’s manuals. All siRNAs, miRNA mimics and NCs were obtained from Biomics Biotechnologies Co., Ltd (China), and the sequences are shown in Table 1.
 |
Table 1 The Sequences of siRNAs and miRNAs |
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from the tissues or cells using TRIzol® reagent (Thermo Fisher Scientific). The mRNA levels of NEK6 were detected using SYBR® Green One-Step qRT-PCR Kit (Thermo Fisher Scientific, #11736-051) according to manufacturer’s protocol, and the reaction system was performed as follows: 5 μL of 2×SYBR Green Reaction Mix, 0.2 μL of SuperScript III RT/Taq Mix, 0.2 μL of forward primer (10 μM), 0.2 μL of reverse primer (10 μM), 1 μL of total RNA, and 3.4 μL of RNase-free dH2O, then subjected to reverse transcription for 5 min at 50 °C and initially denatured at 95 °C for 2 min, and then to 40 cycles of amplification with the condition of 95 °C denature for 15 sec, and 60 °C annealing for 30 sec. The housekeeping gene GAPDH was used as the internal control. The mature miRNA expression levels including miR-26-5p in tissues and cells were detected using miRNA One-step qPCR-PCR SYBR Green kit (Biomics Biotech, China) according to the manufacturer’s protocol, and the reaction system was performed as follows: 5 μL of One-Step SYBR Mix, 0.2 μL of Enzyme Mix, 0.2 μL of forward primer (2 μM), 0.2 μL of reverse primer (5 μM), 0.2 μL of universal reverse primer (URP) (5 μM), 1 μL of RNA template, 3.2 μL of RNase-free dH2O, then subjected to reverse transcription for 30 min at 42 °C and initially denatured at 95 °C for 2 min, and then to 40 cycles of amplification with the condition of 93 °C denature for 10 sec, 60 °C annealing for 30 sec. U6 small nuclear RNA (snRNA) was used as the endogenous internal control. The results were analyzed using the 2−ΔΔCt method.35 The primers were obtained from Biomics Biotechnologies Co., Ltd (China), and the sequences are listed in Table 2.
 |
Table 2 The Primer Sequences of RT-qPCR |
Western Blot Analysis
The cells in logarithmic growth phase were plated into 6-well plates (3×105 cells/well) and continue cultured at 37 °C with 5% CO2 for 24 h. After been treated with siRNAs or miRNAs for 48 h as above, total proteins from cells were lysed in RIPA buffer (Promega) and were quantified using a BCA Protein Assay kit (Beyotime, China). Then, about 40 μg protein per lane was separated using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a PVDF membrane (Sigma-Aldrich), followed by blocking with 5% skimmed milk in Tris Buffer Saline (TBS) buffer (Sangon Biotech, China) for 2 h at room temperature. The blots were detected with the following antibodies: NEK6 (1:500 dilutions; #10378-1-AP), STAT3 (1:1000 dilutions; #10253-2-AP), p-STAT3 (1:1000 dilutions; #39595), Bcl2 (1:2000 dilutions; #68103-1-Ig), Bax (1:2000 dilutions; #50599-2-Ig), MMP-2 (1:500 dilutions; #10373-2-AP), MMP-9 (1:500 dilutions; #10375-2-AP), or GAPDH (1:5000 dilutions; #60004-1-Ig) at 4 °C overnight, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody for 1.5 h at room temperature. All antibodies were purchased from Proteintech (China). ECL Western Blotting Substrate (Thermo Fisher Scientific) was used to visualize the signals and detected by a Chemiluminescence analyzer (ChemiScope 5300 Pro), the mean gray values of blots were quantified by ImageJ version 1.8.0 software (National Institutes of Health).
Immunofluorescence Staining
The cells in logarithmic growth phase were plated onto the small round glass slide in 24-well plates (1×105 cells/well) and kept single-celled suspension, and continue cultured at 37 °C with 5% CO2 for 24 h. Then, the cells were treated with siRNAs or miRNAs for 48 h as above. After being washed in phosphate buffered saline (PBS) (pH7.4) one time, the cells were fixed in 4% paraformaldehyde for 15 min. After being washed in PBS three times for 5 min each time, the cells were then treated with PBS containing 0.1% Triton X-100 at room temperature for 10 min to increase the permeability of cell membrane. The cells were washed in PBS three times for 5 min each time, and incubated in 5% FBS solution at 37 °C for 1 h, and then incubated with primary antibody NEK6 (1:200 dilutions; #10378-1-AP, Proteintech) overnight at 4 °C. After being washed in PBS three times for 5 min each time, the cells were incubated with second antibody in dark at 37 °C for 1 h. The cells were then washed in PBS three times for 5 min each time, and stained with Hoechst 33342 (Thermo Fisher Scientific) in dark at room temperature for 15 min. At last, the cell photos were taken under the fluorescence microscope, and the fluorescence intensity was analyzed with ImageJ software.
Cell Viability Assay
A Cell Counting Kit-8 (CCK-8) assay was used to detect the cell viability. After seeding into 96-well plates (5×103 cells/well) for 24 h, the cells were transfected at 37 °C with si-NEK6, miR-26a-5p mimics or mimics NC for 24, 48 and 72 h. Post 48 h transfection, 10 μL CCK-8 reagent (Beyotime, China) was added into the cells per well and continue incubated at 37 °C for 4 h. The optical density (OD) value of each well was measured using a microplate reader at the wavelength of 450 nm.
Cell Migration Assay
A cell wound healing assay was used to detect the cell migration abilities. Briefly, 5×105 cells which were in logarithmic growth phase were inoculated in a 3.5 cm dish and cultured at 37 °C with 5% CO2 for 24 h. When the cells grown to 90% confluence and covered the bottom of 3.5 cm dish, the cell monolayer scratch was made using 200 μL Pipette Tips. After being washed in PBS, the cells were transfected as above, the cells continue cultured at 37 °C with 5% CO2 for 24 h, 48 h, 72 h and 96 h, the cell migration was observed under the microscope (Olympus). The migration distance was measured by Image J software.
Cell Invasion Assay
A transwell assay was used to detect the cell invasive abilities. In brief, the cells which were treated as above were collected and centrifuged at 1000 rpm for 5 min. After being washed in PBS, the cells were resuspended in medium without FBS, the cell concentration was adjusted to 3×105 cells/mL. The Matrigel (BD Bioscience) was thaw at 4 °C previous day and diluted to 1 mg/mL, and transwells, 24-well plates and Pipette Tips were precooled overnight at −20 °C. Total of 800 μL medium with 10% FBS was added to 24-well plates per well, and the transwells were put into the plates. A 100 μL of Matrigel (1 mg/mL) was added to the upper chamber and incubated for 4 h at 37 °C, and then 200 μL above cell suspension was inoculated into the upper chamber and incubated at 37 °C with 5% CO2 for 24 h. The transwells were got out and washed in PBS, the cells were fixed in 70% pre-cold ethanol, then stained with 0.5% crystal violet solution and incubated at room temperature for 20 min. After being washed with PBS, the cells in front of the upper chamber were wiped using the cotton swab, and at last, the cells were observed under the microscope. The invasive cell numbers were measured using Image J software.
Cell Apoptosis Assay
Flow cytometry based on the Annexin V-FITC/Propidium Iodide (PI) staining was used to detect the cell apoptosis. Post 48 h transfection as above, 1×106 cells were collected and then stained using an Annexin V-FITC Apoptosis Detection kit (Bestbio, China) according to the manufacturer’s protocol. In brief, the cells were resuspended in 1×Binding Buffer after being washed with PBS, and then stained with Annexin V-FITC for 15 min and PI for 10 min at room temperature separately in the dark. Next, the stained cells were analyzed by a flow cytometry (FACSCalibur, BD Biosciences) and BD CellQuest™ Pro software (BD Biosciences).
Dual-Luciferase Reporter (DLR) Assay
The binding site of NEK6 with miR‑26a-5p was displayed using TargetScan, and the binding effects of NEK6 with miR‑26a-5p were validated using DLR assay. Wild-type (WT) or mutated (MUT) 3’UTR of NEK6 mRNA sequence was constructed into a pmirGLO vector (Promega) as luciferase reporter gene vectors. In brief, 2×105 HEK293T cells were seeded into a 24-well plate. After being cultured at 37 °C for 24 h, the cells were co-transfected with pmirGLO-NEK6-3’UTR WT or pmirGLO-NEK6-3’UTR MUT reporter vector and miR-205-3p mimics or mi-NC using Lipofectamine® 2000 (Thermo Fisher Scientific) according to manufacturer’s protocol. At 48 h post-transfection, the Firefly and Renilla luciferase activities were detected using a Dual-Luciferase Reporter Assay System (Promega), the results were normalized to the activities of Renilla luciferase.
Statistical Analysis
The data are presented as the mean value ± standard deviation (SD) and analyzed using SPSS Statistics 19.0 (IBM Corp.) and graphics were made using GraphPad Prism 9.0 (GraphPad Software Inc.). Differences between two groups were performed using unpaired Student’s t-test; differences between multiple groups were analyzed using followed by Dunnett’s post hoc test was used to determine the correlation between NEK6 and miR-26-5p expression was evaluated by Spearman’s rank correlation parametric test. P<0.05 was considered statistically significant.
Results
NEK6 is Highly Expressed in Sarcomas and Osteosarcoma
The expression of NEK6 in human cancers was analyzed using analysis tools of TIMER, UALCNA and GEPIA, and the data were obtained from TCGA databases. TIMER showed that the transcript per million (TPM) values of NEK6 were high in many types of human cancers (Figure 1A). For expression analysis in box plot and survival analyses by UALCNA, NEK6 was overexpressed in sarcoma compared with the normal tissue. Meanwhile, the high expression of NEK6 in the patients with sarcoma had a worse prognostic outcome (Figure 1B and C). Based on the GEPIA analysis, the TPM values of NEK6 also high in most cancers, and NEK6 expression in sarcoma was higher than normal (Figure 1D and E), and its overexpression indicated a shorter overall survival (Figure 1F).
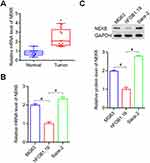
To explore NEK6 expressions in osteosarcoma, NEK6 expression in 13 cases of patients of osteosarcoma and para-carcinoma normal tissues were examined using RT-qPCR. The results showed that NEK6 was overexpressed in 13 OS tissues when compared with normal tissues (Figure 2A). In osteosarcoma cell lines MG63 and Saos-2, the mRNA and protein levels of NEK6 were also highly expressed detected by RT-qPCR and Western blot, respectively, compared with the normal osteoblast cell hFOB1.19 (Figure 2B and C).
The founding indicates NEK6 is a potential oncogene in sarcoma and osteosarcoma and may influence the prognosis and overall survival of patients with sarcoma and osteosarcoma.
NEK6 Was Effective Silenced by siRNAs in Osteosarcoma Cells
To evaluate the role of NEK6 expression in osteosarcoma, the siRNAs were designed and used to downregulate the NEK6 expression on osteosarcoma cells. RT-qPCR result showed that the designed si-NEK6-1, si-NEK6-2 and si-NEK6-3 were all silenced the mRNA levels of NEK6 up to 37%, 14% and 54% in MG63, and 43%, 37% and 60% in Saos-2 cells, compared with si-NC treated cells (Figure 3A and B), and the best silenced siRNA was si-NEK6-3; thus, it was used in the following experiments and labeled as si-NEK6. The protein levels of NEK6 were further investigated in MG63 and Saos-2 cells, the results showed that NEK6 protein levels were inhibited significantly by si-NEK6, compared with untreated cells and si-NC treated cells (Figure 3C).
In addition, the expression of NEK6 was detected using immunofluorescence staining method. The results showed that, the fluorescence intensity of NEK6 expression was lower than that in untreated and si-NC treated cells, both in MG63 and Saos-2 cells (Figure 3D and E).
The results indicate that NEK6 is highly expressed in osteosarcoma cells and can be effectively silenced by NEK6 specific-targeted siRNA.
NEK6 Knockdown Inhibits the Proliferation, Migration, Invasion and Promotes the Apoptosis of Osteosarcoma Cells
To explore NEK6 function in osteosarcoma, the cell proliferation, migration and invasion were detected. To determine cell proliferation following NEK6 knockdown was detected using CCK-8 assay. To compare with the untreated cells and si-NC treated cells, the proliferation abilities of MG63 and Saos-2 were inhibited significantly by si-NEK6 at 48, 72 h and 96 h (Figure 4A and B). The cell migration following NEK6 knockdown was detected using a wound healing assay. To compare with the untreated cells and si-NC treated cells, the migration abilities of MG63 and Saos-2 were inhibited significantly by si-NEK6 (Figure 4C and D). The cell invasion following NEK6 knockdown was detected using a Transwell assay. To compare with the untreated cells and si-NC treated cells, the invasion abilities of MG63 and Saos-2 were inhibited significantly by si-NEK6 (Figure 4E).
On the other hand, the cell apoptosis was measured by FCM based on the Annexin V-FITC and PI staining. The results showed that, the apoptosis of MG63 and Saos-2 cells were promoted by si-NEK6, compared with untreated cells and si-NC treated cells (Figure 4F).
NEK6-Targeted miRNAs Prediction and Validation
Bioinformatics analysis was performed to predict NEK6-targeted miRNAs, which include 28 miRNAs based on TargetScan, 31 miRNAs based on tarbase, 86 miRNAs based on microT-CDS, 150 miRNAs based on Starbase. Through Venn diagrams, 6 miRNAs including miR-141-3p, miR-200a-3p, miR-26a-5p, miR-26b-5p, miR-27a-3p and miR-27b-3p were screened out (Figure 5A). Furthermore, the expression levels of miR-141-3p, miR-200a-3p, miR-26a-5p, miR-26b-5p, miR-27a-3p and miR-27b-3p were detected in osteosarcoma cells, and the result showed that miR-26a-5p levels were lowly expressed in both MG63 and Saos-2 cells, compared with that in hFOB1.19 cells (Figure 5B). Thus, miR-26a-5p was chosen for the following study.
The binding site of miR-26a-5p on NEK6 was displayed by TargetScan and validated by DLR assay. The DLR reporter vectors with the wild-type (WT) and mutant (MUT) 3’UTR of NEK6 mRNA were constructed (pmirGLO-NEK6-3’UTR WT and pmirGLO-NEK6-3’UTR MUT) (Figure 5C). DLR assay showed that transfection of miR-26a-5p mimic decreased the Firefly luciferase activities containing NEK6-3’UTR WT, while no obvious changes on the Firefly luciferase activities containing NEK6-3’UTR MUT were observed (Figure 5D).
These results indicate that miR-26a-5p is lowly expressed in osteosarcoma and NEK6 is the direct target of miR-26a-5p.
miR-26a-5p is Lowly Expressed and Negative Correlation with NEK6 Expression in Osteosarcoma
To investigate miR-26a-5p expression in the patients with osteosarcoma, the expression levels of miR-26a-5p in tumor and para-carcinoma normal tissues were detected by RT-qPCR, and showed that miR-26a-5p levels were significantly lower in tumor tissues than that in normal tissues (Figure 6A). And the correlation between miR-26a-5p and NEK6 expression in osteosarcoma tissues was further analyzed and showed that miR-26a-5p expression and NEK6 expression in osteosarcoma was negative correlation (r=−0.633, P=0.0193) (Figure 6B).
Additionally, to assess the regulation of miR-26a-5p expression in osteosarcoma cells, since miR-26a-5p was lowly expressed osteosarcoma, thus miRNA mimics were used for cell transfection. In MG63 and Saos-2 cells, the levels of miR-26a-5p were obvious up-regulated by miR-26a-5p mimics, compared with untreated and mimic NC treated cells (Figure 6C), and the protein levels of NEK6 were inhibited significantly by miR-26a-5p mimics detected by Western blot assay (Figure 6D).
miR-26a-5p Up-Regulation Inhibits the Proliferation, Migration, Invasion and Promotes the Apoptosis of Osteosarcoma Cells
To explore miR-26a-5p function in osteosarcoma, the cell proliferation, migration and invasion were detected. To compare with the untreated cells and mimics NC treated cells, CCK-8 assay showed that the proliferation abilities of MG63 and Saos-2 were inhibited significantly by miR-26a-5p mimics at 48 h, 72 h and 96 h (Figure 7A and B). Wound healing assay showed that the migration abilities of MG63 and Saos-2 were inhibited significantly by miR-26a-5p mimics (Figure 7C and D). Transwell assay showed that the invasion abilities of MG63 and Saos-2 were inhibited significantly by miR-26a-5p mimics (Figure 7E). FCM based on the Annexin V-FITC and PI staining showed that, the apoptosis of MG63 and Saos-2 cells were promoted by miR-26a-5p mimics (Figure 7F).
NEK6 Inhibited by miR-26a-5p May Be Through STAT3 Signaling Pathway in Osteosarcoma Cells
In normal physiological conditions, STAT3 signaling is strictly regulated to maintain a transient state of activation, and persistent STAT3 activation is found in approximately 70% of human solid tumors and hematocarcinoma.36 It has been suggested that NEK6 may compete with JAK in the STAT3 signaling pathway, and NEK6 recruitment and accumulation may lead to Tyr705 phosphorylation of STAT3, further regulating transcriptional activation of downstream genes.37 The phosphorylation levels of STAT3 were detected to explore the STAT3 signal activation. Besides, STAT3 signal also showed the significant modulation on the genes involved in cell apoptosis and metastasis,38 thus cell apoptosis-related genes Bcl2 and Bax, and metastasis-related genes MMP-2 and MMP-9 were also measured by Western blot. The results showed that phosphorylation levels of STAT3 (p-STAT3) were inhibited by miR-26a-5p mimics significantly, and the expressions of Bax were promoted while Bcl2, MMP-2 and MMP-9 were inhibited by miR-26a-5p mimics in MG63 and Saos-2 cells, compared with the mimics NC treated cells (Figure 8).
The results indicate that NEK6 is an activator of STAT3, metastasis inhibition and apoptosis promotion regulated by miR-26a-5p inhibition of NEK6 may be through STAT3 signaling pathway in osteosarcoma (Figure 9).
 |
Figure 9 Predictive mechanism diagram of NEK6 in osteosarcoma. NEK6 promotes the progression of osteosarcoma through activating STAT3 signaling pathway by down-regulation of miR-26a-5p. |
Discussion
Primary osteosarcoma affects children, adolescents and the elderly, and its age-specific incidence varies according to the histological subtype. Osteosarcoma usually occurs in patients aged 10–30 years, and the proportion varies among different populations,39 and osteosarcoma often occurs in the long bones.40 The causes of osteosarcoma are complex involving genetic mutations and epigenetic pathways.41 The significant genomic complexity and significant interpatient genetic heterogeneity were found in osteosarcoma samples through genome-wide and exome sequencing, transcriptome assessment of gene expression, and epigenetic modification.42
As one of the less studied kinase families, the exact molecular mechanism of NEK family in tumorigenesis remains unclear.17 Recent studies have shown that the NEK family is abnormally expressed in a variety of tumors, such as hepatocellular carcinoma,43 colorectal cancer,44 lung cancer,45 gastric cancer,46 breast cancer,47 prostate cancer48 and pancreatic cancer.49 However, the studies of NEK family are still in the initial discovery stage, and there is currently no in-depth molecular mechanism research. In the study, we found NEK6 was overexpressed in sarcoma and osteosarcoma, and its overexpression was correlated with poor survival of patients. Furthermore, through multifunctional experiments in osteosarcoma cells, we explored that NEK6 was high expressed in osteosarcoma cells, and the cell proliferation, migration and invasion were inhibited by its knockdown, while the cell apoptosis was promoted, indicating NEK6 may be a potential oncogene in sarcoma and osteosarcoma. The results suggest that NEK6 can act as a diagnostic and therapeutic target for osteosarcoma.
In this study, NEK6-targeted miRNAs were predicted by multiple miRNA prediction and analysis software, as well as osteosarcoma cell expression screening and DLR validation were used, showed that miR-26a-5p targeting NEK6 was down-expressed in MG63 and Saos-2 cells also in osteosarcoma patients and negatively correlated with NEK6 expression in osteosarcoma. Furthermore, the expressions of NEK6 could be inhibited efficacy by miR-26a-5p up-regulation in osteosarcoma cells, and resulted in the inhibition of cancer cell proliferation, migration and invasion while the promotion of cell apoptosis. The results suggest that the strategy of NEK6 inhibited by miR-26a-5p may be a perspective method for osteosarcoma treatment.
Signal transducer and activator of transcription factors (STATs) belongs to the family of cytoplasmic transcription factor proteins, responsible for the signal transduction of extracellular cytokines and growth factors and the activation of gene transcription.50 STAT3 is an important member of the STAT family and a downstream oncogenic mediator of JAK-STAT signaling pathway, which promotes cancer cell proliferation and survival. It has been suggested that NEK6 may be a protein kinase of STAT3, a potential kinase substrate molecule for NEK6, and phosphorylation of Tyr705 is responsible for activation of JAK/STAT signaling in the classical JAK/STAT3 pathway.51 Studies have suggested that NEK6 may compete with JAK in the STAT3 signaling pathway, and the recruitment and accumulation of NEK6 may lead to Tyr705 phosphorylation of STAT3,37 thus inhibiting transcriptional activation of downstream apoptosis genes and promoting transcriptional activation of proliferation-related genes.38 Herein, we further explored the STAT3 signaling pathway regulated by miR-26a-5p through NEK6 inhibition, and we found that phosphorylated STAT3 was inhibited by miR-26a-5p significantly, and the expressions of apoptosis-related gene Bcl2 were inhibited and Bax was promoted, while metastasis genes (MMP-2 and MMP-9) were inhibited in osteosarcoma cells.
Conclusion
We first reported that NEK6 was overexpressed in osteosarcoma, and its inhibition resulting in the inhibition of proliferation, migration and invasion, and apoptosis promotion of osteosarcoma cells. Furthermore, NEK6-targeted miR-26a-5p had been found, and its lowly expression in osteosarcoma was negatively correlated with NEK6 expression and biological functions including proliferation, migration, invasion and apoptosis in osteosarcoma cells. However, in the current study, the number of samples of clinical patients is still small and no in-vivo experiments have been carried out. We will further collect more samples and establish animal models to verify our conclusions more comprehensively. The founding suggest that NEK6 is a potential oncogene while miR-26a-5p is a suppressor miRNA in osteosarcoma, and the strategy of NEK6 inhibited by miR-26a-5p through STAT3 signaling pathway may be an effective treatment method and approach of osteosarcoma.
Data Sharing Statement
The underlying data support the results of the present study and can be obtained from the corresponding author upon reasonable request.
Ethical Approval
This study was approved by the Ethics Committee of Affiliated Hospital of Nantong University and followed the guidelines described in the Helsinki Declaration. Written informed consent was obtained for tissue sample collection from each participant.
Funding
This work was supported by the Science and Technology Project of Nantong City (nos. MSZ2022055 and JCZ20121) and the Scientific Research Project of Jiangsu Provincial Commission of Health and Family Planning (no. Y2018099).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Yu L, Zhang J, Li Y. Effects of microenvironment in osteosarcoma on chemoresistance and the promise of immunotherapy as an osteosarcoma therapeutic modality. Front Immunol. 2022;13:871076.
2. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi:10.3322/caac.21654
3. Marchandet L, Lallier M, Charrier C, et al. Mechanisms of resistance to conventional therapies for osteosarcoma. Cancers. 2021;13:683.
4. Lu KH, Lu PW, Lu EW, et al. Curcumin and its analogs and carriers: potential therapeutic strategies for human osteosarcoma. Int J Biol Sci. 2023;19:1241–1265. doi:10.7150/ijbs.80590
5. Eker N, Tokuc AG, Yılmaz B, et al. Outcomes of osteosarcoma in children without high-dose methotrexate: could it be less toxic without effecting survival rates? J Adolesc Young Adult Oncol. 2022;11:252–258.
6. Urlić I, Jovičić MŠ, Ostojić K, et al. Cellular and genetic background of osteosarcoma. Curr Issues Mol Biol. 2023;45:4344–4358.
7. Hız M, Karaismailoglu B, Ulutas S, et al. The effect of preoperative radiotherapy on local control and prognosis in high-grade non-metastatic intramedullary osteosarcoma of the extremities. Arch Orthop Trauma Surg. 2021;141:1083–1089. doi:10.1007/s00402-020-03494-4
8. Fauske L, Jebsen NL, Bondevik H, et al. Exploring the patient perspective of bone sarcoma survivors who have undergone particle radiotherapy abroad. Anticancer Res. 2023;43:2031–2039.
9. Ouyang H, Wang Z. Predictive value of the systemic immune-inflammation index for cancer-specific survival of osteosarcoma in children. Front Public Health. 2022;10:879523. doi:10.3389/fpubh.2022.879523
10. Dong Z, Liao Z, He Y, et al. Advances in the biological functions and mechanisms of miRNAs in the development of osteosarcoma. Technol Cancer Res Treat. 2022;21:1–16.
11. Meltzer PS, Helman LJ. New horizons in the treatment of osteosarcoma. N Engl J Med. 2021;385:2066–2076. doi:10.1056/NEJMra2103423
12. Kelley LM, Schlegel M, Hecker-Nolting S, et al. Pathological fracture and prognosis of high-grade osteosarcoma of the extremities: an analysis of 2847 Consecutive Cooperative Osteosarcoma Study Group (COSS) patients. J Clin Oncol. 2020;38:823–833. doi:10.1200/JCO.19.00827
13. Beird HC, Bielack SS, Flanagan AM, et al. Osteosarcoma. Nat Rev Dis Primers. 2022;8:77.
14. Hattinger CM, Patrizio MP, Fantoni L, et al. Drug resistance in osteosarcoma: emerging biomarkers, therapeutic targets and treatment strategies. Cancers. 2021;13:2878. doi:10.3390/cancers13122878
15. Li YY, Guo L, Han ZM. Roles of NEK family in cell cycle regulation. Yi Chuan. 2021;43:642–653.
16. Moniz L, Dutt P, Haider N, et al. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011;6:18. doi:10.1186/1747-1028-6-18
17. Panchal NK, Evan Prince S. The NEK family of serine/threonine kinases as a biomarker for cancer. Clin Exp Med. 2023;23:17–30. doi:10.1007/s10238-021-00782-0
18. Yu Y, Shen T, Zhong X, et al. NEK6 is an injury-responsive kinase cooperating with STAT3 in regulation of reactive astrogliosis. Glia. 2022;70:273–286. doi:10.1002/glia.24104
19. He Z, Ni X, Xia L, et al. Overexpression of NIMA-related kinase 6 (NEK6) contributes to malignant growth and dismal prognosis in human breast cancer. Pathol Res Pract. 2018;214:1648–1654. doi:10.1016/j.prp.2018.07.030
20. Simabuco FM, Kobarg J, Severino MB. NEK6 regulates redox balance and DNA damage response in DU-145 prostate cancer cells. Cells. 2023;12:256. doi:10.3390/cells12020256
21. Zhang H, Li B. NIMA-related kinase 6 as an effective target inhibits the hepatocarcinogenesis and progression of hepatocellular carcinoma. Heliyon. 2023;9:e15971. doi:10.1016/j.heliyon.2023.e15971
22. Gerçeker E, Boyacıoglu SO, Kasap E, et al. Never in mitosis gene A-related kinase 6 and Aurora kinase A: new gene biomarkers in the conversion from ulcerative colitis to colorectal cancer. Oncol Rep. 2015;34:1905–1914. doi:10.3892/or.2015.4187
23. Orenay-Boyacioglu S, Kasap E, Gerceker E, et al. Expression profiles of histone modification genes in gastric cancer progression. Mol Biol Rep. 2018;45:2275–2282. doi:10.1007/s11033-018-4389-z
24. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14:dmm047662. doi:10.1242/dmm.047662
25. Záveský L, Jandáková E, Weinberger V, et al. Ascites in ovarian cancer: microRNA deregulations and their potential roles in ovarian carcinogenesis. Cancer Biomark. 2022;33:1–16. doi:10.3233/CBM-210219
26. Li PP, Li RG, Huang YQ, et al. LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability. Aging. 2021;13:24171–24191. doi:10.18632/aging.203672
27. Wang Z, Liu T, Xue W, et al. ARNTL2 promotes pancreatic ductal adenocarcinoma progression through TGF/BETA pathway and is regulated by miR-26a-5p. Cell Death Dis. 2020;11:692. doi:10.1038/s41419-020-02839-6
28. Taiwen L, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi:10.1158/0008-5472.CAN-17-0307
29. Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi:10.1016/j.neo.2022.01.001
30. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi:10.1093/nar/gkx247
31. McGeary SE, Lin KS, Shi CY, et al. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:eaav1741. doi:10.1126/science.aav1741
32. Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi:10.1093/nar/gkx1141
33. Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–173. doi:10.1093/nar/gkt393
34. Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi:10.1093/nar/gkt1248
35. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
36. Hashimoto S, Hashimoto A, Muromoto R, et al. Central roles of STAT3-mediated signals in onset and development of cancers: tumorigenesis and immunosurveillance. Cells. 2022;11:2618. doi:10.3390/cells11162618
37. Fu B, Xue W, Zhang H, et al. MicroRNA-325-3p facilitates immune escape of mycobacterium tuberculosis through targeting LNX1 via NEK6 accumulation to promote anti-apoptotic STAT3 signaling. mBio. 2020;11:e00557–20. doi:10.1128/mBio.00557-20
38. Shao M, Lou D, Yang J, et al. Curcumin and wikstroflavone B, a new biflavonoid isolated from Wikstroemia indica, synergistically suppress the proliferation and metastasis of nasopharyngeal carcinoma cells via blocking FAK/STAT3 signaling pathway. Phytomedicine. 2020;79:153341. doi:10.1016/j.phymed.2020.153341
39. Beauchemin MP, Raghunathan RR, Accordino MK, et al. New persistent opioid use among adolescents and young adults with sarcoma. Cancer. 2022;128:2777–2785. doi:10.1002/cncr.34238
40. Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi:10.1155/2011/548151
41. Todosenko N, Khlusov I, Yurova K, et al. Signal pathways and microRNAs in osteosarcoma growth and the dual role of mesenchymal stem cells in oncogenesis. Int J Mol Sci. 2023;24:8993. doi:10.3390/ijms24108993
42. Sayles LC, Breese MR, Koehne AL, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9:46–63. doi:10.1158/2159-8290.CD-17-1152
43. Zhang Y, Wang W, Wang Y, et al. NEK2 promotes hepatocellular carcinoma migration and invasion through modulation of the epithelial-mesenchymal transition. Oncol Rep. 2018;39:1023–1033. doi:10.3892/or.2018.6224
44. Sabir SR, Sahota NK, Jones GD, et al. Loss of Nek11 prevents G2/M arrest and promotes cell death in HCT116 colorectal cancer cells exposed to therapeutic DNA damaging agents. PLoS One. 2015;10:e0140975. doi:10.1371/journal.pone.0140975
45. Shi YX, Yin JY, Shen Y, et al. Genome-scale analysis identifies NEK2, DLGAP5 and ECT2 as promising diagnostic and prognostic biomarkers in human lung cancer. Sci Rep. 2017;7:8072. doi:10.1038/s41598-017-08615-5
46. Cao Y, Song J, Chen J, et al. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine. 2018;97:e9630. doi:10.1097/MD.0000000000009630
47. Anuraga G, Wang WJ, Phan NN, et al. Potential prognostic biomarkers of NIMA (never in mitosis, gene A)-related kinase (NEK) family members in breast cancer. J Pers Med. 2021;11:1089. doi:10.3390/jpm11111089
48. Singh V, Jaiswal PK, Ghosh I, et al. The TLK1-Nek1 axis promotes prostate cancer progression. Cancer Lett. 2019;453:131–141. doi:10.1016/j.canlet.2019.03.041
49. Yan Z, Qu J, Li Z, et al. NEK7 promotes pancreatic cancer progression and its expression is correlated with poor prognosis. Front Oncol. 2021;11:705797. doi:10.3389/fonc.2021.705797
50. La Manna S, De Benedictis I, Marasco D. Proteomimetics of natural regulators of JAK-STAT pathway: novel therapeutic perspectives. Front Mol Biosci. 2022;8:792546. doi:10.3389/fmolb.2021.792546
51. McMurray JS, Mandal PK, Liao WS, et al. The consequences of selective inhibition of signal transducer and activator of transcription 3 (STAT3) tyrosine705 phosphorylation by phosphopeptide mimetic prodrugs targeting the Src homology 2 (SH2) domain. JAKSTAT. 2012;1:263–347. doi:10.4161/jkst.22682
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.