Back to Archived Journals » Nanobiosensors in Disease Diagnosis » Volume 4
Nanoscale virus biosensors: state of the art
Authors Krejcova L, Michalek P, Merlos Rodrigo M, Heger Z , Krizkova S, Vaculovicova M, Hynek D, Adam V , Kizek R
Received 15 March 2015
Accepted for publication 15 May 2015
Published 6 August 2015 Volume 2015:4 Pages 47—66
DOI https://doi.org/10.2147/NDD.S56771
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Chulhee Choi
Ludmila Krejcova,1 Petr Michalek,2 Miguel Merlos Rodrigo,2 Zbynek Heger,2 Sona Krizkova,1 Marketa Vaculovicova,1 David Hynek,1 Vojtech Adam,1 Rene Kizek1
1Central European Institute of Technology, Brno University of Technology, 2Department of Chemistry and Biochemistry, Faculty of Agronomy, Mendel University in Brno, Brno, Czech Republic
Abstract: Since the beginning of the new millennium, viruses have shown huge epidemiological and pandemic potential: severe acute respiratory syndrome (SARS) in 2002, pandemic swine flu in 2009, and last but not least the West African Ebola outbreak in 2014. The occurrence and spread of the new virus in pandemic dimension poses a threat to the health and lives of 7 billion people worldwide. There is a growing urgency for highly sensitive and selective detection techniques, usable for a wide number of applications, including disease diagnosis, pharmaceutical research, agriculture, as well as preventive measures. Nanobiosensors represent a new promising tool for virus detection. This review gives a brief survey of the issue of viral detection, comprising diagnostics of target structure of viruses such as nucleic acids or proteins. This review covers different detection principles, methods of fabrication, and applications of virus biosensors.
Keywords: nanoparticle, nanomaterial, diagnostics, immunosensors, hybridization
Introduction
The research field of biosensors began with the introduction of the glucose oxidase biosensor in 1962, introduced by Clark and Lyons.1 Since then many interesting sensor and biosensor applications have been described, and some of them have been commercialized. The most widely accepted definition of a biosensor is: “an analytical device which includes a biologically active element (or components) in close contact with an appropriate physicochemical transducer to generate a measurable signal directly proportional to the concentration of target substance(s) in the sample”.2–4 A typical biosensor consists of three parts: a biological recognition component (enzyme, antibody, DNA, etc), a sensor element for signal acquisition (electrical, optical, or thermal), and an element for amplification/signal processing.5 A scheme of a typical biosensor is shown in Figure 1. A way of signal conversion depends on the type of physicochemical change resulting from the initial and final signal.6 The most frequently used biological component of sensors are enzymes,7–10 antibodies,8,11 and oligonucleotides.12–14 Therefore, the biosensor differs from the sensor by the presence of biological (biorecognition) component, which usually exhibits a bioaffinity or biocatalytic role. The principle of biocatalytic role is the conversion of the analyte during the chemical reaction.15,16 In case of the bioaffinity role, the analyte is bound specifically and selectively to the biorecognition element.
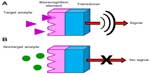  | Figure 1 Schematic diagram of biosensor with target analyte (A) and nontarget analyte (B). |
Infectious diseases are the main cause of significant increase in pathogenesis and death throughout the world, surpassing even the cardiovascular diseases and cancer.17 In developed countries, a remarkable technological progress in sanitation to identify and control most of the infectious diseases has been achieved.18 Biosensors combine the sensitivity of detection methodologies and constitutional selectivity of biomolecules.6,17 Efforts to develop highly sensitive, fast, stable, and low-cost biosensors have been enabled by extensive and arduous research.6
Viral cultivation is considered as the gold standard for virus detection. Viral isolation techniques are accurate and sensitive, but getting results take as long as several days.19–21 Some viruses are hard or impossible to cultivate. In these cases, fertilized chicken embryos or experimental animals are used for virus cultivation.22–24 More recent techniques, such as polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA), can directly detect pathogen-specific DNA/RNA or proteins. When we compared these techniques, PCR is more sensitive than ELISA. However, both of them have some disadvantages, which are hidden in strict laboratory conditions, requiring well-trained staff, expensive instruments, and time-consuming processes.19,25–27 Therefore, the development of rapid, accurate, dependable, and miniaturized devices for virus detection is still needed.28
The glucose biosensor was the first and the most successful bisensor, based on technology which was developed by Clark and Lyons1 more than 40 years ago. If these biosensors play such an important role, could biosensors for virus detection acquire similar position? What is hidden in the future? Which type of biorecognition molecule and which technology is suitable for the rapid and sensitive detection of viral diseases? The purpose of a viral biosensor is to provide real-time diagnosis, far as possible in the early stage of infection. If therapy is initiated in the early stage of infection, it significantly increases the success rate of the selected treatment.29
There is no doubt that the future seems promising for biosensing technology. This fact has been demonstrated by the increasing number of articles with the topics: “biosensor and virus” published on Web of Science (in 2000, 57 publications; in 2005, 117 publications; in 2010, 200 publications, and in the year 2014, 211 publications; overall 2,457 articles). The number of papers published/patents issued per year is an important indicator of research activity, and the present growth rate of over 16% suggests that the future looks bright indeed.30 But on the other hand, there is only one truly commercially successful biosensor, and it is suggested that between 80% and 90% of research activity in this area rarely results in a commercial product.31 However, the growth in biosensor research indicates the higher likelihood of another successful biosensor in the future, which looks positive despite very little progress over the past several years.
Biorecognition molecules for virus biosensors
Biosensors should offer rapid, highly specific and sensitive, fast detection of viral diseases.32 Crucial in design and biosensor function is improvement of the affinity, selectivity, and specificity, which could determine the success or failure of the whole detection technology. So, it is difficult to estimate which biorecognition element to use for a given target pathogen.19,32 There are two main biorecognition strategies: detection of viral nucleic acid (NA) sequence32–34 and detection of specific viral biomolecules such as surface proteins/antigens.35–37 Nanotechnology-based biosensors show high specificity and sensitivity after labeling with NA probe, antibody, or other specific molecule with affinity to the target structure.38
Nucleic acids
NA-based biosensors have actually been a hot topic and hold huge promise for clinical diagnosis.39 The past two decades have evidenced the development of various NA biosensors based on different detection methods, including optical,40,41 electrochemical,4,32,42 electrochemiluminescence (ECL),43,44 quartz crystal microbalance (QCM),45,46 and surface plasmon resonance (SPR) techniques.47,48 In general, NA-based detection is more specific and sensitive than immunological-based detection, while the second one is faster and more robust.25 For signal generation or amplification, the probe can be labeled with a variety of labeling molecules such as electroactive substances, fluorophores, radioisotopes, enzymes, or, more recently, haptens (to which antibodies are available).32,49 Hybridization biosensors have potential to obtain higher sensitivity and selectivity than conventional methods. The optimal hybridization efficiency can be achieved by control of the distribution and orientation of probes on the transducer surface.39 Nowadays, application of nanomaterials for fabrication of transducers has become more and more popular and widespread. Quantum dots, nanotubes, nanowires, nanoparticles, magnetic particles, and, more recently, nanopillars are the most attractive signal transducers.50,51 The applications of nanotechnology, with unique properties to construct novel biosensors, are constantly being expanded upon by researchs.4,38
Antigens/specific proteins/receptors
Viral infections are often associated with the presence of generic, not specific, symptoms, and thus their origin is hardly diagnosed.52,53 The presence of the specific antibodies or antigens enables the detection of specific viral pathogen and enables to start the appropriate treatment. Antibodies are one of the most frequently used biorecognition elements for biosensor fabrication54–57 and are produced by the host in response to the presence of foreign molecules and organisms.52 The antibody-based diagnostics market is still growing, and therefore a new, rapid, and accurate immunodiagnostic method is required. To date, several strategies for “reagentless biosensors” based on antibodies and natural or engineered binding proteins have been described.54 In the past, polyclonal antibodies were used first; recently, they were pushed out by high-affinity monoclonal antibodies. For research purposes, monoclonal or polyclonal antibodies can be raised specifically against a protein, another antibody, or even a whole virus and can bind with high affinity (Kd=106−109 M).58 Peptides (polymeric amino acids) can specifically bind viral proteins or antibodies, too. These short peptides can be designed and synthesized by phage display.59−61 Phages with strong cognitive abilities are used for fabrication of biosensors. Another way of virus detection is based on glycan–protein interactions, which are of high importance in several biological processes. A variety of carbohydrate residues, associated as the “glycol-codes”, provide the fundamental keys for specific and high affinity “lock-in” recognition events associated with a wide range of pathologies.62 A very new approach offers the design and synthesis of artificial receptors (molecularly imprinted receptors). They are able to recognize and bind different target molecules with high affinity and specificity comparable to their biological counterparts, provide a number of advantages such as greater long-term storage stability, potential reusability, resistance to microbial spoilage, and custom synthesis without the need to inoculate laboratory animals, as well as facile integration with transducers.63 Karimian et al64 suggested that synthetic receptors are able to successfully rebind the template with exceptional rebinding properties.
Detection methods of virus biosensors
Recent advances in virus biosensing have been made, especially in fluorescence,65 light scattering,66,67 surface-enhanced Raman scattering (SERS),68,69 electrochemical,7,34,70 QCM,71,72 microcantilevers (MCLs),73,74 and SPR17,47 sensors. The breakdown structure of “nanobiosensor for viral detection” segmentation is described in Figure 2. The objective of the search for a new biosensor is to invent an alternative tool for effective viral disease diagnosis via a compact format that is not time consuming and does not require highly trained personnel or enhanced laboratory facilities.
  | Figure 2 Structure of “nanobiosensor for virus detection” segmentation. |
Electrochemical biosensors
Electrochemical biosensors (EBs) have shown a great success in recent years because of the unique properties and easy-to-use platform for a wide range of practical applications in the fields of medical diagnostics, clinical genetic analysis, forensic analysis, and environmental monitoring.75–77 Electrode transducers are often used for virus detection due to their easily modifiable surface and compatibility with microfabricated technology.78 The sensitive layer is represented by an interface between the working electrode surface and the analyzed environment. The cornerstone of EB is the immobilization of the biorecognition element (receptor) on the electrode surface. As a receptor, a probe (hybridization EB) or a targeting molecule (affinity EB) is commonly used. These two basic strategies common for electrochemical biosensing are illustrated in Figure 3. The electrode (sensor) can be modified by probe or antibody (recognition element) (Figure 3B), followed by target isolation (Figure 3C), and terminated by simple signal detection or detection after signal amplification (Figure 3D). The complex formed from the receptor and the target molecule at the electrode surface results in a detectable change of the signal and is converted into a quantitative amperometric,79 potentiometric,80,81 or impedimetric signal.38 A wide range of electrochemical methods can be used for detection include differential pulse voltammetry (DPV),35,80,82 square wave voltammetry (SWV),83,84 cyclic voltammetry (CV),85–87 and conductometry.88,89 Also, impedance is widely used as a detection method in biosensors.11,38,90,91 To enhance the signal intensity, labels could be utilized. In general, two methods in the design of EB assays have been exploited, including label and label-free approaches.32 Electrical techniques do not strictly require the attachment of labels, but these are usually less sensitive.19
Nucleic acid electrochemical biosensors
Nucleic acid electrochemical biosensors (NAEB) have been pushed to the forefront due to high sensitivity, specificity, portability, and integrated compatibilities with microelectronics.92 The potential application of the NAEB in gene analysis, diagnosis, environmental and food safety monitoring were reported by different authors.92 The transduction is more direct than for other techniques because the biochemical process is directly transduced into an electrical response.34 The most severe limitation of NAEB is the detection limit. A number of scientists consider fluorometric assays much more sensitive.19,34 On the other hand, some recent studies focusing on the detection limit of NAEB reported analysis of NA at femtomolar93,94 and attomolar levels.95
The immobilization of the oligonucleotide probes on the surface of the electrode is a key step to fabricate the electrochemical oligonucleotide biosensor. It is not surprising that various electrodes [carbon, mercury, and gold] have been modified and tested for NA biosensing.4,96,97 DNA, peptide nucleic acid (PNA), or locked nucleic acid (LNA) probes can be grafted onto a solid substrate where direct hybridization with an unlabeled DNA target occurs. Electrochemical transduction during hybridization was matched with or without a redox indicator.34,95,98 Numerous strategies have been developed to increase their sensitivity, selectivity, and speed.
The NAEB could be classified into three groups, based on the strategies of the bioelectrochemical transduction, as follows:
- direct change of electroactivity after hybridization,
- change of signal after solid-surface immobilization,
- amplification of signal change using active labels.
Change of direct electroactivity after hybridization
The first strategy is based on direct change of electroactivity caused by the oxidation/reduction of guanine, cytosine, and/or adenine after hybridization reaction.42,99 In this case, a signal change could be detected directly without necessity of electroactive labels. In the text, we describe only pioneering, interesting, or outstanding articles.
Zari et al100 described human papilloma virus (HPV) biosensor, where unlabeled DNA probes have been immobilized by coadsorption of thiolated oligonucleotides onto the sensing surface of a screen-printed gold electrode. After hybridization, DNA is treated with an acid, and the acid-released purine bases are directly determined by SWV with HPV sequence detection limit of 2 pg·mL−1(S/N =3).100 The specific sequence for hepatitis C virus-1 (HCV-1) was detected by Pournaghi-Azar et al,101 using a label-free DNA hybridization biosensor. The sensor relied on the immobilization of a 20-mer oligonucleotide (containing 2 guanines and 11 cytosines) as a probe on the pencil graphite electrode. The hybridization event was monitored by DPV of the guanine signal.101 Huang et al102 described a label-free EB with dual amplification strategy based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzymes nanowires. Electrochemical signals were obtained by measuring the increase in reduction current of oxidized tetramethylbenzidine sulfate, which was generated by DNAzyme in the presence of H2O2. This method exhibited ultrahigh sensitivity toward avian influenza A (H7N9) virus DNA sequence, with detection limits of 9.4 fM.102 Another EB was fabricated by conjugation of a biotinylated probe DNA and an avidin-modified glassy carbon electrode to detect the influenza virus.87 The current value of the biosensor was evaluated after hybridization of the probe and target DNA using CV, and hybridization was reflected by decrease in the current value.87
Change of signal intensity after solid-surface immobilization
The second strategy included EB, where electroactivity of nucleotides immobilized on the solid support is amplified. Different authors employed a variety of nanomaterials such as graphene,103,104 gold nanoparticles (AuNPs),102,105 magnetic and nonmagnetic particles,106,107 and carbon nanotubes108 for modification of the base of the transducers.
Chitosan/Fe3O4 nanobiocomposite-based platform for electrochemical detection of human immunodeficiency virus 1 (HIV-1) was described by Lam Dai et al.32 The sensitivity of nanoparticles was enhanced using methylene blue (MB), and SWV method and SPE were used for measurement.32 Diamond is another extremely attractive material, used as a biosensing interface. Synthetically prepared diamonds have outstanding electrochemical properties such as inertness and biocompatibility. Qureshi et al7 described a novel biosensing platform based on geometrically controlled DNA bonding using diamond nanowires, followed by the electrochemical sensing. A biosensor based on an anion exchange nanoporous membrane under direct current bias was described by Senapati et al.109 The ionic diode feature is associated with external surface-charge inversion on the positively charged nanomembrane upon hybridization of negatively charged target on the probes on the membrane.109 The resulting bipolar membrane can be used to accurately quantify the hybridization reaction between the probe and the target sequences of dengue virus (DV). The limit of detection was 1 pM for 27 base sequence in a 15 minute assay.109
Gao et al110 described label-free detection by applying rolling-circle amplification (RCA) based on silicon nanowire field-effect transistor. The probe was immobilized on the silicon nanowire transistor surface, followed by sandwich hybridization of the target DNA and RCA primer that acted as a primer to hybridize the RCA template, which created a long ssDNA product and enhanced the electronic responses.110
Shi et al111 described a hybrid microarray, realized by a facile template-free method on gold substrates. The formation mechanism was based on self-assembled monolayers (SAMs) between gold substrates and hybrid crystals.111 A highly oriented hybrid microarray was formed on the vertical SAMs. The previously described label-free EB was used for the detection of the avian influenza virus (AIV) H5N1. A nanoporous alumina membrane-based EB, where DNA probes were immobilized onto the alumina channel walls, was proposed by Rai et al.112 Alumina nanoporous membrane-like structure was carved over a platinum wire electrode by electrochemical anodization, and the subsequent probe–target hybridization inside the pores influenced the pore size and ionic conductivity.112
Amplification of signal change using active labels
The third strategy is based on electroactive labels or intercalators for amplification of target/probe NA signal.
An ECL hybridization biosensing system for the detection of HIV-1 gene, based on a super-sandwich ds DNA probe and ruthenium complex as an intercalated signal-producing compound, was described by Ruan et al.113 First, the probe was self-assembled on the gold electrode, then target HIV-1 gene was hybridized, and thereafter two auxiliary probes were hybridized to form the supersandwich. Finally, the ECL indicator (ruthenium complex) was intercalated into the supersandwich.113 Lin et al described EB based on the horseradish peroxidase (HRP)- and exonuclease III-assisted target recycling amplification strategy. Another strategy was described by Lin et al,93 a molecular beacon probe with hairpin structure was assembled on the electrode and labeled by HRP to give a strong initial signal. Upon target DNA sensing, the probe was removed by the exonuclease III accompanied by the release of the target for the hybridization and cleavage. Biotin-labeled mononucleotides were liberated, biotin and HRP binding on the electrode decreased as did the HRP-amplified electrochemical current. This dual enzyme strategy provided an ultrasensitive approach with detection limit at 10 fM.93 Malecka et al43 described a genosensor based on the ion-channel mimetic mechanism, where the signal generated upon hybridization was recorded by a redox-active marker [Fe(CN)6]3−/4− using voltammetric techniques. An EB based on immobilized anthraquinone (AQ)-labeled pyrrolidinyl PNA probe was developed for the selective detection of HPV by Jampasa et al.114 The redox-active label AQ was covalently attached to the probe, which was immobilized onto carbon SPE. Target DNA hybridization (AQ signal response) was measured using SWV.114 Ahour et al115 described an assay for detection of double-stranded plasmid without denaturation, using PNA as a probe. The gold electrode was modified with 6-mercapto-1-hexanol, following self-assembly of the monolayer of cysteine conjugated with PNA probe, which binds dsP of HCV. The signal response of PNA/double-stranded plasmid triplex formation was mediated by MB and measured by DPV with detection limit of 9.5 pg·μL−1.115
An oligonucleotide-incorporated, nonfouling surface was constructed to resist nonspecific absorption by Liu et al.116 Fully matched target DNA templated the ligation between a probe and a tandem signal of HRP and generated amperometric signal.116 Qi et al117 described EB, where carbon ionic liquid electrode (CILE) was used as the basal electrode. The Co3O4 nanorods (nano-Co3O4), graphene, and chitosan were mixed together to form a nanocomposite material and casted on the CILE surface.117 Using MB as the electrochemical indicator, the hybridization reactions were monitored with the reduction peak current.117
Chen et al118 described EB for detection of HIV based on a cascade hybridization of the capture probe, target, and two auxiliary probes and formation of micrometer-long one-dimensional DNA nanostructures. To target nanostructure formation and signal amplification, redox indicator [Ru(NH3)6]3+ was used.118
Immunoelectrochemical biosensors and protein affinity EB
Electrochemical immunosensors could potentially replace routinely used ELISA for diagnosis of viral diseases. The advantage of an electrochemical immunosensor is the direct detection of the antigen–antibody complex formed on a surface layer.119 Recent trends in development of electrochemical immunosensors are focused on the new transducers, which are able to improve immobilization of antibodies, sensitivity, dynamic range of detection, and attempts to regenerate the sensor surface.119
Different nanostructures and electroactive labels have been used to improve the sensitivity of methods.120,121 Alipour et al121 described a capacitive-based immunobiosensor for detection of the hepatitis B surface antigen, where AuNPs were attached to a secondary antibody in order to improve the sensitivity of the method. Due to the relatively large size of the particles and the thickness of the dielectric layer, the capacitance changed remarkably. Therefore, the detection limit was improved to about 10 ng mL−1.121 Miodek et al122 described the electrochemical immunosensor for PB1-F2 influenza protein detection based on an integrated ferrocenyl group involved as a redox marker for signal detection. The proposed biosensor and specific anti-PB1-F2 monoclonal antibody could be applied for studying PB1-F2 during influenza infection.122 Miodek et al123 also described PB1-F2 EB based on immunodetection of the PB1-F2 oligomerization. The immunosensor was based on conductive polypyrrole modified with ferrocenyl groups as a redox marker for enhancing signal detection. Antibodies specific for monomeric or oligomeric PB1-F2 forms were immobilized on polypyrrole matrix via biotin/streptavidin layer.123
In the work of Hong et al,124 concanavalin A was placed on a nanostructured gold electrode, which selectively captured noroviruses. Secondary antibodies conjugated with alkaline phosphatase were used to improve the signal. CV revealed a linear relationship (R2=0.998) between current and concentration of noroviruses, with a relatively short assay time (1 hour) and a good detection limit (35 copies·mL−1).
An electrochemical immunosensor based on gold-film electrode, obtained from a recordable compact disk (CD-electrode), was developed by Cavalcanti et al125 for nonstructural protein 1 (NS1) of DV detection. Anti-NS1 monoclonal antibodies were immobilized on the CD-electrode via protein A.125 The NS1 interaction with anti-NS1 immobilized on CD-electrode was detected by DPV.125 The immunosensor showed the detection limit of 0.33 ng mL−1.125 Silva et al86 constructed thiophene-modified SPE also for detection of the DV NS1, an important marker for acute-phase diagnosis. The thiophene SPE was coated with AuNPs conjugated to Protein A, the anti-NS1 antibodies were immobilized via their Fc portions via Protein A.86 Amperometric responses to the NS1 of DV were obtained by CV in the presence of ferrocyanide/ferricyanide as redox probe.86 Xie et al126 reported EB for AIV H5 subtype detection using graphene oxide-H5-polyclonal antibodies-bovine serum albumin (GO-PAb-BSA) nanocomposite. The graphene oxide (GO) carrying H5-polyclonal antibody was used for signal amplification. This immunosensor showed a 256-fold increase in detection sensitivity compared to the immunosensor without GO-PAb-BSA.126
Braustein and Braustein127 reported a novel EB for detection of the bacteriophage virus MS2, using nanoporous oxirane-derivatized beads. These beads are commercially evaluated for bioconjugation of antibodies, enabling detection of a viral concentration as low as 10 PFU.mL−1, where PFU stands for plaque-forming units.127 Immunoamperometric techniques, using a commercial kit, were used to validate the accuracy of novel technology for virus concentration determination.127 Huang et al128 reported a polysilicon nanowire-based biosensor system-on-chip, fabricated by complementary metal-oxide-semiconductor (CMOS) process. In addition, an on–off key wireless transceiver was also integrated to form a wireless biosensor system-on-chip technology.128 This was a pioneering work to harness the momentum of CMOS-integrated technology into emerging biodiagnosis technologies, and examined to have label-free and low-concentration biomolecular detection, as a consequence. Such developed technology can be a promising candidate for on-field and personalized applications in biomedical diagnosis.128 Fabrication of biocompatible nanofibrous materials by electrospinning was described by Luo et al.129 The novel nanostructure improves the biochemical binding effect and sensor signal-to-noise ratio. Luo et al129 presented the electrospinning method of nitrocellulose nanofibrous membrane and its antibody functionalization for bovine viral diarrhea virus detection. The antibody attachment and pathogen-binding effect were verified using the confocal laser scanning microscope and scanning electronic microscope.129 With the advantage of efficient antibody functionalization, excellent capillary capability, and relatively low cost, the electrospinning process and surface functionalization can be implemented to produce nanofibrous membranes for different immunodetection.129
Electrochemical impedance biosensors
Electrochemical impedance spectroscopy (EIS) was exploited in the detection of a number of viruses such as HIV, DV, influenza virus, herpes virus, hepatitis C and B viruses.130–136 Also EIS can be involved in biosensing of viral pathogens as hybridization immunobiosensors.132,137,138 The conventional EIS is mainly based on the Faradaic process where charge is transferred across the interface.134 Different redox-systems, most frequently [Fe(CN)6]4−/3−, are added to the electrolyte. Rickert et al139 observed that the long-term presence of the redox-system ([Fe(CN)6]4−/3−) reduces the activity of the protein layer. An immunosensor based on non-Faradaic process, a nonlabeling methodology, has also been researched and has proved to be somewhat more accessible to point-of-care applications.139,140
Electrochemical impedance biosensors (EIBs) are typically constructed on a SAM layer141,142 or on a conductive polymer layer.143,144 The process of hybridization or antibody–antigen interaction is measurable by response in conductivity across the immunosensor surface, which is translated into a change in the resistance and/or double-layer capacitance following analyte capture.145 Detection of the change in capacitance is easier to measure as no reference electrode is required and therefore it is more durable “in-field”. EIS method can exhibit much higher sensitivity than other conventional counterparts such as amperometric, voltammetric, and potentiometric measurements.146
Hybridization impedance biosensor
Mashhadizadeh and Talemi147 described a hepatitis B virus (HBV) EB based on covalent immobilization of the probe on the AuNPs, which were functionalized onto a gold electrode. The DNA biosensor fabrication was characterized by CV and EIS using [Fe(CN)6]3−/4−, and the probe–target hybridization process was observed by DPV.147 An EIB for the detection of AIV H5N1 specific sequence has been proposed by Malecka et al.137 The NH2-ssDNA probe was deposited onto a gold electrode surface.137 The genosensor was capable of determining complementary sequences, with the detection limit in the fM range.137 Aydinlik et al13 introduced EIS hybridization biosensor for detection of influenza B virus. The detection method utilized AuNPs and Meldola’s Blue as an intercalator label on a pencil graphite electrode.13 The hybridization process was confirmed with EIS and CV.13 A novel and integrated membrane sensing platform for detection of specific sequence of DV, based on an anodic aluminum oxide membrane was described by Deng and Toh.48 Platinum electrodes (50–100 nm thick) were coated directly by the alumina membrane to eliminate the solution resistance outside the nanopores.48 The EIS was employed to monitor the impedance changes within the nanopores upon the DNA binding and showed the pore resistance increases linearly in response to the increasing concentration of the target DNA in the range of 1×10−12−1×10−6 M.48
Connection between hybridization- and immuno-EIB was reported by John et al138 as a detection of specific sequence and glycoprotein (gp120) of HIV virus, based on the novel generation 4 poly(propylene imine) dendrimer/streptavidin platform. The biosensor was prepared by the immobilization of probe and aptamer on the modified electrode to detect complementary DNA and gp120, respectively. Responses were evaluated with EIS and SWV in the presence of [Fe(CN)6]3−/4−.138
EIS immunobiosensor
The interdigitated electrodes and electrical measurements for the diagnosis of dengue infection using antigen–antibody conjugation method were reported by Fang et al.134 Preinactivated DV was immobilized indirectly onto the immunosensor surface, precoated with sol–gel thin film modified with organic SAM and a cross-linker over the interdigitated electrodes.134 The modified sensor surface served as a selective sensing probe to capture/conjugate the dengue antibody molecules.134 Hnaien et al135 described an immunological sensor based on functionalized gold electrode allowing for the detection of rabies antigen. This biosensor is based on the immobilization of antirabies antibodies onto functionalized gold microelectrode and the antibody–antigen interaction.135 Tung et al148 described the detection of weak molecular binding between the DV and its receptor C-type lectin domain family 5, member A (CLEC 5A), which is critical for DV-induced hemorrhagic fever and DV pathogenesis. Through a highly sensitive nanostructured sensing electrode of AuNPs deposited on a nanohemisphere array, a label-free detection of the ultraweak binding between CLEC 5A and the DV can be performed with EIS. Jiang and Spencer132 described cell biosensor based on EIS for the precise counting of human CD4+ helper lymphocytes. In this biosensor, the sensing area was composed of densely packed working electrode pixels, each of which was comparable to a single CD4+ cell in size, thus enabling diagnose HIV.132 CD4+ helper cells were captured on the chemically modified electrode pixels and detected individually by monitoring the interfacial impedance changes on each independent pixel.132 Mishra et al149 fabricated a miniaturized microelectrode on-chip detection method to quantify human CD4+ cells through impedance measurements made with simple and battery operated electronics in a handheld device. The microelectrode was modified with protein G, human albumin, monoclonal mouse antihuman CD4, and mouse immunoglobulin.149 The CD4+ cells present in human blood were verified by impedance and CV.149
Many EIS detection systems for various viruses have been described, but most of them were dedicated to influenza detection, based on utilization of anti-influenza antibody or aptamers. Kiilerich-Pedersen et al150 reported a study where influenza A (H1N1) virus aptamers were linked covalently to the conductive polymer microelectrodes. H1N1 virions were captured by immobilized probes, and detected as changes in the impedance. Nidzworski et al11 described an assay based on the direct attachment of antibodies to the gold electrode, which allows detection of the influenza virus with sensitivity similar to molecular methods. Jarocka et al151 described an immunosensor for the avian influenza hemagglutinin (HA) H5, where gold electrode was modified by gold colloidal nanoparticles functionalized by antibody-binding fragments of anti-H5 monoclonal antibodies. The antigen–antibody interaction was explored with EIS in the presence of [Fe(CN)6]3−/4−.151 The immunosensor was able to recognize three different His-tagged variants of recombinant HA (H5N1) with detection limit of 2.2 pg mL−1.151 Wang et al90 described the impedance biosensor for detection of H5N2 influenza virus-infected chickens, based on a combination of magnetic nanobeads coated with the subtype-specific antibody for the capture of the target virus, and the microfluidic chip with an interdigitated array microelectrode for impedance detection of virus complexes. Lin et al91 described an immunobiosensor for H5N1 and H5N2 which could work stand-alone or be connected with a laptop via USB (universal serial bus) interface. Results showed that this impedance biosensor could identify H5N1 virus with a detection limit of 103 EID50 mL−1 in 30 minutes, where EID50 stands for 50% egg infectious dose.91 Wang et al152 described an EIB for subtype H5N1 detection based on an interdigitated array, and it is described in detail in Figure 4. Polyclonal antibodies against H5N1 influenza surface antigen HA were functionalized on the gold microelectrode (Figure 4A); thereafter, target H5N1 viruses were captured (Figure 4B), resulting in a change in the impedance of the interdigitated array microelectrode surface.152 Red blood cells were used for amplification (Figure 4C) of the antibody–antigen reaction.152 The binding of target H5N1 onto the antibody-modified microelectrode was confirmed by atomic force microscopy.152
Quartz crystal microbalance
The basic part is the AT-cut quartz crystal (where the quartz blank is in the form of thin plate cut at an angle of about 35°15′ to the optic axis of the crystal) that exhibits piezoelectric behavior. The quartz crystal’s frequency depends on the speed of the shear wave and the thickness of the crystal. Maximum sensitivity to the difference of the mass is in the center of the crystal.153 In 1959, Sauerbrey described the relationship between frequency decrease (Δf) and deposition of mass (m) on the crystal surface in the air (vacuum).154–156 The QCM frequency change is also dependent on the density and viscosity of the solution, where a liquid is passed over the quartz crystal.157 An electrical field applied to the QCM produces mechanical stress that induces an acoustic wave to travel in a direction perpendicular to the surface of the crystal.66
QCM is a rapid, easy to use, and relatively simple method that can be applied in real-time detection of mass changes on the crystal surface, such as hybridization and/or antigen–antibody reactions.146,158 Receptors for viral target substances (probes, antibodies) are capable of binding to the terminal active functional groups of SAMs and capturing targets.159,160 The QCM can consequently detect mass changes due to these molecular interactions on the surface of the QCM.66
During the last decade, a new approach in the form of functionalized nanoparticles has been exploited to increase the specificity and sensitivity of QCM biosensors.161 QCM biosensing can be applied in various fields of biotechnology, such as clinical diagnosis159,162 and detection of the occurrence of epidemiologically important diseases.159
Hybridization QCM biosensor
Skladal et al158 reported the use of QCM to detect HCV DNA in serum using biotinylated DNA probes, immobilized onto a biotin-tagged SAM surface via streptavidin coupling. Zhou et al163 developed a piezoelectric HBV DNA biosensor, where HBV probe was immobilized onto the gold electrode with polyethyleneimine adhesion, glutaraldehyde cross-linking method, and physical adsorption method. This sensor represents a rapid, sensitive, and reliable alternative to the common HBV DNA determination.163 Dell’Atti et al164 used QCM in combination with PCR for HPV detection. Target was detected using DNA probes immobilized onto a SAM layer.164 The study of Hong et al165 described a rapid and sensitive QCM biosensor for diagnosis of viral hemorrhagic septicemia infection in fish. The main viral RNA-encoding G protein was detected by the specific probe.165 Three different probes and three different immobilization methods were employed.165 The most efficient (avidin–biotin probe immobilization) was more sensitive and much faster than a conventional reverse transcription-PCR, with detection limit of 0.0016 μM.165
Yao et al166 described the application of RCA-based QCM for the detection of HBV sequence. After amplification, the RCA product is maintained during the assay through the covalent bonding between the capture probes and the gold electrode surface.166 Using high amplification efficiency of Phi29 DNA polymerase, results show that RCA has significantly enhanced sensitivity for the target.166
QCM immunobiosensor and affinity QCM biosensor
Wang and Li71 reported QCM aptasensor based on ssDNA cross-linked polymeric hydrogel for detection of AIV H5N1. The aptamer and H5N1 virus binding results in abrupt swelling of the hydrogel, and this was monitored by frequency decrease.71 In comparison with the anti-H5 antibody immunosensor, the hydrogel QCM aptasensor lowered the detection limit and reduced the detection time.71 Owen et al167 developed and characterized QCM system for the direct detection of aerosolized influenza A virions. SAM of mercaptoundecanoic acid was formed on QCM gold electrodes for the immobilization of anti-influenza A antibodies for capture of influenza virions. The proposed biosensor can play an important role in the public health by offering a new analytical tool for identification of biocontaminated areas.167 Hewa et al168 described a QCM biosensor for the detection of both influenza A and B viruses in clinical samples. Conjugation of AuNPs to the detecting antibody improved the mass sensitivity of the immunosensor and showed that QCM techniques were comparable in sensitivity and specificity to cell culture methods.168 Wangchareansak et al169 reported a study where N-acetylglucosamine was immobilized as the ligand on the gold surface of a QCM via the S–H bond, after which wheat germ agglutinin was used to mimic the real target (influenza HA). Lu et al170 developed a biomimetic sensor for the detection of HIV type 1 related protein (glycoprotein 41, gp41) based on an epitope imprinting technique. Dopamine was used as the functional monomer and polymerized on the surface of quartz crystal in the presence of template, a synthetic peptide – analogous to residues 579–613 of the gp41 for the specific target binding.170 Chen et al171 proposed the formation of molecularly imprinted membrane for the epitope (NS1) of DV on the gold QCM electrode and developed an immunosensor for the virus detection. The response of QCM to NSl showed a comparable frequency shift to those chips immobilized with monoclonal antibodies, and authors also demonstrated a method for antibody detection by forming a sandwich.171 Liu et al172 described an immunosensor employing conducting polymer entrapment method to immobilize immunoprotein on the QCM for clinical flow injection analysis. An immunoassay of anti-pseudorabies virus antibody in mouse sera further exemplified its practical potential in diagnostic implication.172 Lee and Chang160 reported flow type of QCM for the real-time determination of bovine ephemeral fever virus, SAM was used for the immobilization of the bovine ephemeral fever virus monoclonal antibody on the gold surface of QCM; thereafter, positive correlation was found between the virus concentration and frequency changes (R2=0.9962) on this QCM system.160
Optical biosensors
Recent advances in biosensor technologies have potential to deliver point-of-care diagnostics of diverse sensing strategies including optical biosensor.173 Detection of viruses is essential for pharmaceutical industry, disease prognosis, and surveillance. Optical techniques are very sensitive, and can detect even single molecule, but require the attachment of a fluorophore molecule to the target.174,175
Optical coherence tomography
Optical coherence tomography (OCT) is a recently developed imaging technique for studying the rheological properties of tissues in vivo.176 This technique offers noninvasive imaging in real time with high resolution and is potentially suitable for monitoring of tissues as well as biosensor fabrication.176,177 The OCT has helped to usher in a new era of in vivo diagnostic imaging.178 Trantum et al179 reported a biosensor that used secondary flows arising from surface Marangoni stresses that are necessary to produce signal in the proposed design. These evaporation-driven flows generate signal in the assay based on a polydimethylsiloxane substrate but not substrates with greater thermal conductivity like indium tin oxide-coated glass. In this proof-of-concept design, the M13K07 bacteriophage was used as a model target.179
Lee et al180 described a unique system integrating several optoelectronic-based biological diagnostic tools such as an ellipsometer, a laser Doppler vibrometer/interferometer, SPR analyzer, an interference microscope, a photon tunneling microscope, an optical coherence tomography unit, and a confocal scanning microscope. This OBMorph system, useful as a powerful optical metrology diagnostic tool, can be used at the beginning of sensor chip fabrication, during the signal detection/monitoring, and in the final biological analysis.180
Optical fluorescence
The availability of highly sensitive and selective fluorescent labeling techniques makes fluorescence a widely used optical method in microfluidic systems for detection of pathogenic organisms, hormones, or other medically relevant analytes.181
Kim et al182 developed a double-stranded and dual-anchored aptamer on reduced GO nanosheets for speedy and specific detection of the target protein in biological and clinical patient samples. As a model target protein, interferon-gamma was used. This approach allowed a rapid quantification of the target protein in HIV-positive serum samples.182 Another option for virus identification by optical fluorescence detection is the application of the retroviruses tagged with a genetically encoded pH-sensor and a fluorescent content marker enabled simultaneous measurements of the pH drop within virus-carrying vesicles and the resulting virus–endosome fusion.183 Label-free chemiresistive sensors based on a polypyrrole nanoribbon were batch-fabricated by a lithographically patterned nanowire electrodeposition technique. A plant-pathogen-specific antibody was covalently conjugated on the surface of the structure via N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide/N-hydrosuccinimide cross-linking. The sensing performance was investigated by the detection of cucumber mosaic virus and showed excellent sensitivity.28 A new method for improving the sensitivity for detection of the bacteriophage virus MS2 using thin films of nanoporous silicon was developed. A viral concentration was detectable by measuring the fluorescence from the exposed porous silicon film.184 Iyer et al41 showed engineered nanoscale ZnO nanostructures acting as an efficient platform for enhancing fluorescence detection capacity toward sensing cDNA without the need for amplification. Such an inexpensive and rapidly synthesized ZnO platform developed by ultrasonic spray pyrolysis was used for the first time in enhanced fluorescence detection of all of four serotypes of DV labeled with four different fluorophores in one single detection system.
Optical light scattering
For few decades, light scattering techniques have been powerful, though difficult to use tools. Grepstad et al185 designed a biomolecule antigen/specific protein sensor that uses cross-polarized excitation and detection for increased sensitivity. The sensor can be made both cheap and compact to facilitate use at point-of-care.185 A local evanescent, array-coupled biosensor was used to detect spherical polystyrene nanoparticles with diameters of 40 and 200 nm, whose sizes and refractive index are similar to virus particles. This detection of virus-like nanoparticles via scattering using a chip-scale optical biosensor showed high effectivity. Mie scattering in an evanescent field theory was used to model the scattered light intensity for both sizes of nanoparticles.186 The utilization of microparticle immunoagglutination assays using forward light scattering measurements in a microfluidic chip was used for detecting viral particles. The model target was bovine viral diarrhea virus. In the microfluidic chip, the virus was detected in low concentration, down to a concentration of 103 TCID50 mL−1.187 Vesicular stomatitis rhabdovirus was used as a generic model for capture, detection, and identification of a number of pathogenic viruses by field modulated light scattering.188
For NA detection, Lu et al67 showed a gold nanorods-based biosensor for the detection of HBV DNA based on fluorescence resonance energy transfer. The biosensor exhibited good selectivity, even for single-mismatched DNA detection.67 Strong et al188 showed how field modulated light scattering can be employed in a label-free assay to identify and quantify a broad range of targeted microbial species using affinity probes and can also be used for sequence specific detection of amplified DNA for HIV-1.
Surface-enhanced Raman scattering
The SERS method has a great potential for the detection of Raman-active species. The application of the SERS method for the detection of single molecules, biomolecules, and even cells has increased dramatically over the past few years. A SERS using silver nanorod array substrates had developed, allowing for rapid detection of trace levels of viruses with a high degree of sensitivity and specificity.189 Researchers have investigated the formation of hot spots, which are small regions of a highly enhanced electromagnetic field that indicate high SERS intensity.190 In addition to its high sensitivity, the SERS method has several other advantages, including the ability to fingerprint individual molecules, narrower spectral peaks compared to fluorescence peaks, a single excitation source, minimal photobleaching, and low background from aqueous environments.191 These features make SERS a perfect tool for the development of diagnostic assays.
For antigen/specific protein detection, SERS assay can detect spectral differences between viruses, viral strains, and viruses with gene deletions in biological media.192 The method provides rapid diagnostics for detection and characterization of viruses generating reproducible spectra without viral manipulation.193 Lin et al194 described the focused ion beam technique, which was employed to precisely fabricate hexagon-like Au nanorods arrays as a surface enhanced Raman scattering active substrate. A “ring diameter” was created by the convergence of three hexagon-like Au nanorods with respect to the dimension of the target viruses, such as adenovirus, encephalomyocarditis virus, and influenza virus (H1N1) with different sizes.194 This scattering biosensor provided good discrimination ability for distinguishing viruses of various sizes or virus strains.194 Also, inverted triangular Au nanocavities with various indentation depths and tip-to-tip displacements were well arrayed as a substrate for qualitative virus detection.195 Through the induction of the electromagnetic effect by the substrate, the virus can be distinguished from the amino acids on its surface. The detectable concentration for encephalomyocarditis virus or adenovirus was 106 PFU.mL−1 and for influenza virus was 104 PFU.mL−1.195 A highly sensitive immunoassay utilizing SERS has been developed with a new Raman reporter and a unique SERS-active substrate incorporated into a microfluidic device for detection of HBV antigen from human blood.196 Figure 5 shows a SERS biosensor for antigen or specific protein detection, consisting of three steps: SERS substrate modification by antitarget antibody (Figure 5A), target isolation, followed by SERS tag (Figure 5B) binding, and SERS tag detection (Figure 5C).
For NA detection, Lim et al197 showed that DNA on AuNPs facilitates the formation of well-defined gold nanobridged gap particles that generate a highly stable and reproducible SERS signal. Label-free optical detection of viral nucleoprotein binding to a polyvalent anti-influenza aptamer by monitoring the SERS spectra of the aptamer–nucleoprotein complex was a novel method for identification of influenza viruses.198 Pang et al199 developed a simple and sensitive assay for the detection of the RNA genetic marker associated with highly pathogenic influenza virus by SERS.
SPR biosensors
The SPR-based biosensing is one of the most advanced label-free, real-time detection technologies.200 SPR is a collective oscillation of free charges present at the interface of two media (metal–dielectric), with permittivities of opposite sign.201 This method is based on measuring the refractive index of very thin layers of material adsorbed on a metal surface. At certain conditions, surface plasmons on a metallic film can be excited by photons, transforming a photon into a surface plasmon depending on the refractive index of the adsorbate.202 A wide range of reaction rates and binding affinities can be measured using dynamic or steady-state analysis.200 Determination of affinity parameters for biomolecular interactions is the most common application of SPR sensors, where antibody–antigen, ligand–receptor, and protein–NA interactions are most often used. But also DNA–DNA or enzyme–substrate interactions can be studied.203 This method can be used for a wide range of applications in microbiology and virology.202
Hybridization SPR biosensor
The NA probe is attached to the surface of the sensor via a functional group attached to one of oligonucleotide ends. Three most widely used approaches are the adsorption using streptavidin–biotin interactions,200,204 thiolate–gold bond,204–206 and covalent bond via terminal amine207 or carboxyl groups208 and maleimide-ethylene glycol bond.209 For the proper functioning of the biosensor, it is crucial to avoid probe hybridization on regions that may affect the binding site of interest and form the secondary structures such as hairpins or loops.210
NA probes can be used for a wide range of biomolecules, including various types of NAs itself. Jin et al211 used SPR for detection of 20 bp oligonucleotides and 405 bp PCR products with concentration ranging from 50 to 400 ng mL−1 and 5 to 60 ng mL−1, respectively. p53 cDNA was determined in a study by Yao et al212 using a carboxylated dextran film immobilized onto the SPR sensor surface. MicroRNA study by Sipova et al213 presented thiol-derivatized DNA probes attached to the surface of gold layer on the chip. NA probes were also used for genotyping of 24 types of HPV.214 Kim et al215 designed SPR biosensor to detect avian influenza NA. Hybridization reactions between target DNA probes and probes immobilized on a gold surface using thiol-modified oligonucleotides were monitored quantitatively by measuring the resonance wavelength in the visible waveband.215 Teng et al216 designed a biotinylated DNA probe immobilized on the SPR chip modified with streptavidin. The prepared biosensor exhibited good sensitivity with a detection limit of 0.5 pM and was able to discriminate perfect complementary and possible mismatches in sequence.216 Not only NAs, but also proteins, transcription factors217–219, and drugs220 were determined using SPR via their interaction with NA. Fisher et al221 tested recombinant HIV-1 nucleocapsid protein as a target to very short biotinylated oligonucleotides. Results showed that even sequences shorter than 10 bases are sufficient for stable binding of molecules.221 NA binding properties of the core protein of HCV were investigated by Baltzinger et al,222 using labeled biotin-NA immobilized on streptavidin-coated CM4 sensor chips. A method of RNA hybridization on the surface of the streptavidin-coated chip to study RNA–protein interactions was presented in a paper by Yang et al.223 Nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus showed a high binding affinity for the leader sequence of viral genome.223
SPR immunobiosensor
Antibodies represent the most common standard for functionalization of the surface of SPR biosensors. In the study of Xu et al,224 an SPR biosensor chip linked with antirabies virus antibodies was developed for the prompt monitoring of the antigens of rabies. N protein-specific antibody and N protein-specific antibody with G protein-specific antibody of rabies virus (RABV) were linked on two different flow cells on one CM5 chip. This chip was tested for the detection of RABV antigens using the crude extract of RABV from infected BHK cells.224 Kumbhat et al225 studied the presence of DV-specific IgM antibodies in dengue-positive sera using covalently immobilized DV antigen on a gold sensor chip. For HBV sensing, gold-binding polypeptide fused with single-chain antibody against HBV surface antigen was developed by Zheng et al.226 An article by Vaisocherova et al227 described the direct, label-free detection of antibodies against the Epstein–Barr virus, with detecting limit of 0.2 ng mL−1.
Affinity and aptamer SPR biosensor
Biotin-labeled aptamers attached to SPR chip coated with streptavidin were used for specific binding of influenza HA in studies by Wang et al228 and Bai et al,229 when aptamer was applied for the detection of AIV H5N1. The specificity was confirmed by comparison of AIV H5N1 with other nontarget AIV subtypes, which showed no interference. Mandenius et al230 evaluated sialic acid-based structures and lectins as ligands for human influenza HA detection. These ligands can be used for the development of a rapid bioanalytical sensor. Suenaga et al231 developed an SPR-based method for analyzing the glycan–HA interactions, using chemically synthesized biotinylated multivalent glycans.
Microcantilevers
MCL-based systems play a significant role in the field of biosensors for the detection of ultrasmall masses such as proteins and other biomolecules because of their small size, light weight, high surface-to-volume ratio, and possible multiplex application.57,232 Cantilever-based sensing involves the transduction of a biomolecular interaction to a measurable mechanical change.233 While the cantilevers do not possess their inherent selectivity for chemical and biological agents, moieties for specific binding features have to be used for coating according to the final application.234 The optimum transducer response is created when the target reacts specifically with only one side of the cantilever, the sensing surface. Immobilization of the moieties to the opposite side of the cantilever has to be minimal as should nonspecific binding of the target on the surface.235
Hybridization MCL biosensors
Method developed by Fritz et al236 proved that a single base mismatch between two 12-mer oligonucleotides is clearly detectable using hybridization of complementary oligonucleotides. Su et al237 applied gold–thiol covalent bonding for DNA strand linkage. Such a modified cantilever was dipped into the target DNA solution for hybridization. Then, AuNP-labeled DNA strands were hybridized on the other end of target DNA.237 Nanoparticles then acted as a nucleating agent for the growth of silver after exposure to photographic developing solution.237 Increased silver mass led to frequency shift, which could be measured. Detectable DNA concentration using this method was lower than 0.05 nM.237
MCL immunobiosensors
Antibodies are useful tools for surface functionalization of cantilevers. For such sensors, the antibody layer represents the sensing element, while the microcantilever acts as a mechanical transducer.235 H1N1 HA peptide was detected using this approach by Bajwa et al.238 Similarly, AIV H9 detection used covalently immobilized monoclonal antibodies, with wide linear response in the 7.6 ng mL−1 to 76 μg mL−1 concentration range, and the detection limit was 1.9 ng mL−1.239 Fritz et al236 also studied the specific binding of the constant region of immunoglobulins to protein A. Distinct differential signals from immunoglobulins, coming from various animals, reflected the known specific binding properties of protein A to immunoglobulins of different mammals.
Conclusion
Viral diseases are one of major threats to health and life of the world population. Therefore, development of rapid and high-sensitivity assays for viral disease detection has tremendous importance for medical healthcare. Current diagnostics methods are pushed out by nanobiosensors at the research stage, and they begin to penetrate medical praxis. Upgraded, more sensitive, more accurate, rapid, and user-friendly viral disease biosensors are still required. The attention of scientists also points to nanoparticles and nanomaterials as a new alternative to biosensor fabrication. Recent advances in biosensors have been focused on the development of miniaturized biosensors with high sensitivity, specificity, and stability. The possibility for commercialization is crucial for the development of biosensors and their transfer into reality.
Acknowledgment
Financial support from the projects CEITECCZ.1.05/1.1.00/ 02.0068 is gratefully acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
References
Clark LC, Lyons C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann N Y Acad Sci. 1963;102(1):29–45. | |
Arnold MA, Meyerhoff ME. Recent advances in the development and analytical applications of biosensing probes. CRC Crit Rev Anal Chem. 1988;20(3):149–196. | |
Wilson GS, Gifford R. Biosensors for real-time in vivo measurements. Biosens Bioelectron. 2005;20(12):2388–2403. | |
Krejcova L, Hynek D, Adam V, Hubalek J, Kizek R. Electrochemical sensors and biosensors for influenza detection. Int J Electrochem Sci. 2012;7(11):10779–10801. | |
Velasco-Garcia MN, Mottram T. Biosensor technology addressing agricultural problems. Biosystems Eng. 2003;84(1):1–12. | |
Singh R, Das Mukherjee M, Sumana G, Gupta RK, Sood S, Malhotra BD. Biosensors for pathogen detection: a smart approach towards clinical diagnosis. Sens Actuator B Chem. 2014;197:385–404. | |
Qureshi A, Kang WP, Davidson JL, Gurbuz Y. Review on carbon- derived, solid-state, micro and nano sensors for electrochemical sensing applications. Diam Relat Mat. 2009;18(12):1401–1420. | |
Cosnier S, Mailley P. Recent advances in DNA sensors. Analyst. 2008;133(8):984–991. | |
Dey RS, Raj CR. Enzyme-integrated cholesterol biosensing scaffold based on in situ synthesized reduced graphene oxide and dendritic Pd nanostructure. Biosens Bioelectron. 2014;62:357–364. | |
Newman JD, Turner AP. Home blood glucose biosensors: a commercial perspective. Biosens Bioelectron. 2005;20(12):2435–2453. | |
Nidzworski D, Pranszke P, Grudniewska M, Krol E, Gromadzka B. Universal biosensor for detection of influenza virus. Biosens Bioelectron. 2014;59:239–242. | |
Muti M, Kuralay F, Erdem A, Abaci S, Yumak T, Sinag A. Tin oxide nanoparticles-polymer modified single-use sensors for electrochemical monitoring of label-free DNA hybridization. Talanta. 2010;82(5):1680–1686. | |
Aydinlik S, Ozkan-Ariksoysal D, Kara P, Sayiner AA, Ozsoz M. A nucleic acid-based electrochemical biosensor for the detection of influenza B virus from PCR samples using gold nanoparticle-adsorbed disposable graphite electrode and Meldola’s blue as an intercalator. Anal Meth. 2011;3(7):1607–1614. | |
Grabowska I, Stachyra A, Gora-Sochacka A, et al. DNA probe modified with 3-iron bis(dicarbollide) for electrochemical determination of DNA sequence of Avian Influenza Virus H5N1. Biosens Bioelectron. 2014;51:170–176. | |
Xu XD, Lin BB, Feng J, et al. Biological glucose metabolism regulated peptide self-assembly as a simple visual biosensor for glucose detection. Macromol Rapid Commun. 2012;33(5):426–431. | |
Gramsbergen JB, Leegsma-Vogt G, Venema K, Noraberg J, Korf J. Quantitative on-line monitoring of hippocampus glucose and lactate metabolism in organotypic cultures using biosensor technology. J Neurochem. 2003;85(2):399–408. | |
Inci F, Tokel O, Wang SQ, et al. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano. 2013;7(6):4733–4745. | |
Carrascosa LG, Moreno M, Alvarez M, Lechuga LM. Nanomechanical biosensors: a new sensing tool. Trac-Trends Anal Chem. 2006;25(3):196–206. | |
Esfandyarpour R, Esfandyarpour H, Harris JS, Davis RW. Simulation and fabrication of a new novel 3D injectable biosensor for high throughput genomics and proteomics in a lab-on-a-chip device. Nanotechnology. 2013;24(46):465301. | |
Hu AZ, Colella M, Tam JS, Rappaport R, Cheng SM. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real-time PCR. J Clin Microbiol. 2003;41(1):149–154. | |
Gueudin M, Vabret A, Petitjean J, Gouarin S, Brouard J, Freymuth F. Quantitation of respiratory syncytial virus RNA in nasal aspirates of children by real-time RT-PCR assay. J Virol Meth. 2003;109(1):39–45. | |
Camargos MF, Rajao DS, Leite RC, Stancek D, Heinemann MB, Reis JK. Genetic variation of bovine leukemia virus (BLV) after replication in cell culture and experimental animals. Genet Mol Res. 2014;13(1):1717–1723. | |
Atkins GJ, Balluz IM, Glasgow GM, et al. Analysis of the molecular-basis of neuropathogenesis of RNA viruses in experimental-animals – relevance for human-disease. Neuropathol Appl Neurobiol. 1994;20(2):91–102. | |
Wang Y-z, Wei J-b, Zhang G-q, Wang W-w, Ma Y, Shi Y-z. Adaptive passage of rabies virus in chicken embryo fibroblasts. Chin J Biol. 2012;25(6):669–671. | |
Qasim M, Lim DJ, Park H, Na D. Nanotechnology for diagnosis and treatment of infectious diseases. J Nanosci Nanotechnol. 2014;14(10):7374–7387. | |
Sun W, Jiao K, Zhang SS. Electrochemical ELISA for the detection of cucumber mosaic virus using o-pheneylenediamine as substrate. Talanta. 2001;55(6):1211–1218. | |
Cella LN, Chen W, Myung NV, Mulchandani A. Single-walled carbon nanotube-based chemiresistive affinity biosensors for small molecules: ultrasensitive glucose detection. J Am Chem Soc. 2010;132(14):5024. | |
Chartuprayoon N, Rheem Y, Ng JCK, Nam J, Chen W, Myung NV. Polypyrrole nanoribbon based chemiresistive immunosensors for viral plant pathogen detection. Anal Meth. 2013;5(14):3497–3502. | |
Oh DY, Barr IG, Hurt AC. A novel video tracking method to evaluate the effect of influenza infection and antiviral treatment on ferret activity. Plos One. 2015;10(3). | |
Parkinson G, Pejcic B. Using Biosensors to Detect Emerging Infectious Diseases. Perth, Australia: Nanochemistry Research Institute, Curtin University of Technology Perth; 2005. | |
Fuji-Keizai USA. Biosensor Market, R&D Applications and Commercial Implication: WS and worldwide. New York, NY: Fuji-Keizai USA; 2004. | |
Lam Dai T, Binh Hai N, Nguyen Van H, Hoang Vinh T, Huy Le N, Phuc Xuan N. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen printed electrodes. Mater Sci Eng C. 2011;31(2):477–485. | |
Solanki PR, Patel MK, Kaushik A, Pandey MK, Kotnala RK, Malhotra BD. Sol-gel derived nanostructured metal oxide platform for bacterial detection. Electroanalysis. 2011;23(11):2699–2708. | |
Lazerges M, Bedioui F. Analysis of the evolution of the detection limits of electrochemical DNA biosensors. Anal Bioanal Chem. 2013;405(11):3705–3714. | |
Krejcova L, Nejdl L, Hynek D, et al. Beads-based electrochemical assay for the detection of influenza hemagglutinin labeled with CdTe quantum dots. Molecules. 2013;18(12):15573–15586. | |
Esseghaier C, Ng A, Zourob M. A novel and rapid assay for HIV-1 protease detection using magnetic bead mediation. Biosens Bioelectron. 2013;41:335–341. | |
Lin Y-Y, Wang J, Liu G, Wu H, Wai CM, Lin Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens Bioelectron. 2008;23(11):1659–1665. | |
Yao CY, Fu WL. Biosensors for hepatitis B virus detection. World J Gastroenterol. 2014;20(35):12485–12492. | |
Gao WH, Zhang A, Chen YS, et al. A novel probe density controllable electrochemiluminescence biosensor for ultra-sensitive detection of Hg2+ based on DNA hybridization optimization with gold nanoparticles array patterned self-assembly platform. Biosens Bioelectron. 2013;49:139–145. | |
Jenison R, Rihanek M, Polisky B. Use of a thin film biosensor for rapid visual detection of PCR products in a multiplex format. Biosens Bioelectron. 2001;16(9–12):757–763. | |
Iyer MA, Oza G, Velumani S, et al. Scanning fluorescence-based ultrasensitive detection of dengue viral DNA on ZnO thin films. Sens Actuator B Chem. 2014;202:1338–1348. | |
Palecek E, Bartosik M. Electrochemistry of nucleic acids. Chem Rev. 2012;112(6):3427–3481. | |
Malecka K, Michalczuk L, Radecka H, Radecki J. Ion-channel genosensor for the detection of specific DNA sequences derived from plum pox virus in plant extracts. Sensors. 2014;14(10):18611–18624. | |
Zaytseva NV, Montagna RA, Lee EM, Baeumner AJ. Multi-analyte single-membrane biosensor for the serotype-specific detection of Dengue virus. Anal Bioanal Chem. 2004;380(1):46–53. | |
Kumar H, Rani R. Development of biosensors for the detection of biological warfare agents: its issues and challenges. Sci Prog. 2013;96(3):294–308. | |
Castro ACH, Franca EG, de Paula LF, et al. Preparation of genosensor for detection of specific DNA sequence of the hepatitis B virus. Appl Surf Sci. 2014;314:273–279. | |
Riedel T, Rodriguez-Emmenegger C, de los Santos Pereira A, et al. Diagnosis of Epstein-Barr virus infection in clinical serum samples by an SPR biosensor assay. Biosens Bioelectron. 2014;55:278–284. | |
Deng JJ, Toh CS. Impedimetric DNA biosensor based on a nanoporous alumina membrane for the detection of the specific oligonucleotide sequence of Dengue virus. Sensors. 2013;13(6):7774–7785. | |
Chan YY, Chua AL, Lim BH, Pattabiraman L, Ravichandran M, inventors; UNIV SAINS MALAYSIA USM (UYSA-Non-standard), assignee. Lateral flow device for detecting target nucleic acid sequence, comprises a conjugates region containing a detector conjugate which binds to a hapten label of the target nucleic acid sequence and is provided with a signaling label. United States patent US WO2011112068-A1. September 15, 2011. | |
Kugel V, Ji HF. Nanopillars for sensing. J Nanosci Nanotechnol. 2014;14(9):6469–6477. | |
Sharma S, Madou M. A new approach to gas sensing with nanotechnology. Philos Trans A Math Phys Eng Sci. 2012;370(1967):2448–2473. | |
Caygill RL, Blair GE, Millner PA. A review on viral biosensors to detect human pathogens. Anal Chim Acta. 2010;681(1–2):8–15. | |
Kirsch J, Siltanen C, Zhou Q, Revzin A, Simonian A. Biosensor technology: recent advances in threat agent detection and medicine. Chem Soc Rev. 2013;42(22):8733–8768. | |
Ueda H, Dong J. From fluorescence polarization to Quenchbody: recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. Biochim Biophys Acta. 2014;1844(11):1951–1959. | |
Urrego LF, Lopez DI, Ramirez KA, Ramirez C, Osma JF. Biomicrosystem design and fabrication for the human papilloma virus 16 detection. Sens Actuator B Chem. 2015;207:97–104. | |
Dover JE, Hwang GM, Mullen EH, Prorok BC, Suh S-J. Recent advances in peptide probe-based biosensors for detection of infectious agents. J Microbiol Meth. 2009;78(1):10–19. | |
Buchapudi KR, Huang X, Yang X, Ji H-F, Thundat T. Microcantilever biosensors for chemicals and bioorganisms. Analyst. 2011;136(8):1539–1556. | |
Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23(9):1126–1136. | |
Wang SQ, Humphreys ES, Chung SY, et al. Peptides with selective affinity for carbon nanotubes. Nat Mater. 2003;2(3):196–200. | |
Su W, Cho M, Nam J-D, Choe W-S, Lee Y. Highly sensitive electrochemical lead ion sensor harnessing peptide probe molecules on porous gold electrodes. Biosens Bioelectron. 2013;48:263–269. | |
Pavan S, Berti F. Short peptides as biosensor transducers. Anal Bioanal Chem. 2012;402(10):3055–3070. | |
Chabre YM, Roy R. Multivalent glycoconjugate syntheses and applications using aromatic scaffolds. Chem Soc Rev. 2013;42(11):4657–4708. | |
Whitcombe MJ. Molecularly imprinted polymers smart hydrogel crystal gardens. Nat Chem. 2011;3(9):657–658. | |
Karimian N, Turner APF, Tiwari A. Electrochemical evaluation of troponin T imprinted polymer receptor. Biosens Bioelectron. 2014;59:160–165. | |
Mukundan H, Anderson AS, Grace WK, et al. Waveguide-based biosensors for pathogen detection. Sensors. 2009;9(7):5783–5809. | |
Wang XH, Li YA, Wang HF, et al. Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens Bioelectron. 2010;26(2):404–410. | |
Lu XC, Dong X, Zhang KY, Han XW, Fang X, Zhang YZ. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst. 2013;138(2):642–650. | |
Tripp RA, Dluhy RA, Zhao YP. Novel nanostructures for SERS biosensing. Nano Today. 2008;3(3–4):31–37. | |
Li M, Cushing SK, Liang HY, Suri S, Ma DL, Wu NQ. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal Chem. 2013;85(4):2072–2078. | |
Iost RM, Madurro JM, Brito-Madurro AG, Nantes IL, Caseli L, Crespilho FN. Strategies of nano-manipulation for application in electrochemical biosensors. Int J Electrochem Sci. 2011;6(7):2965–2997. | |
Wang RH, Li YB. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens Bioelectron. 2013;42:148–155. | |
Dultsev FN, Tronin AV. Rapid sensing of hepatitis B virus using QCM in the thickness shear mode. Sens Actuators B Chem. 2015;216:1–5. | |
Timurdogan E, Alaca BE, Kavakli IH, Urey H. MEMS biosensor for detection of hepatitis A and C viruses in serum. Biosens Bioelectron. 2011;28(1):189–194. | |
He JH, Sun SQ, Ye JS, Lim TM. Self-assembly carbon nanotubes on cantilever biosensor for sensitivity enhancement. Int Mems Conf. 2006;34:423–428. | |
Li F, Feng Y, Dong PJ, Tang B. Gold nanoparticles modified electrode via a mercapto-diazoaminobenzene monolayer and its development in DNA electrochemical biosensor. Biosens Bioelectron. 2010; 25(9):2084–2088. | |
Luo X, Lee TM-H, Hsing IM. Immobilization-free sequence-specific electrochemical detection of DNA using ferrocene-labeled peptide nucleic acid. Anal Chem. 2008;80(19):7341–7346. | |
Adam V, Huska D, Hubalek J, Kizek R. Easy to use and rapid isolation and detection of a viral nucleic acid by using paramagnetic microparticles and carbon nanotubes-based screen-printed electrodes. Microfluidics Nanofluidics. 2010;8(3):329–339. | |
Xue D, Elliott CM, Gong P, Grainger DW, Bignozzi CA, Caramori S. Indirect electrochemical sensing of DNA hybridization based on the catalytic oxidation of cobalt (II). J Am Chem Soc. 2007;129(7):1854. | |
Uliana CV, Tognolli JO, Yamanaka H. Application of factorial design experiments to the development of a disposable amperometric DNA biosensor. Electroanalysis. 2011;23(11):2607–2615. | |
Zhu X, Ai S, Chen Q, Yin H, Xu J. Label-free electrochemical detection of avian influenza virus genotype utilizing multi-walled carbon nanotubes-cobalt phthalocyanine-PAMAM nanocomposite modified glassy carbon electrode. Electrochem Commun. 2009;11(7):1543–1546. | |
Liu F, Choi KS, Park TJ, Lee SY, Seo TS. Graphene-based electrochemical biosensor for pathogenic virus detection. BioChip J. 2011;5(2):123–128. | |
Liu YT, Deng J, Xiao XL, et al. Electrochemical sensor based on a poly(para-aminobenzoic acid) film modified glassy carbon electrode for the determination of melamine in milk. Electrochim Acta. 2011;56(12):4595–4602. | |
Krejcova L, Huska D, Hynek D, et al. Using of paramagnetic microparticles and quantum dots for isolation and electrochemical detection of influenza viruses’ specific nucleic acids. Int J Electrochem Sci. 2013;8(1):689–702. | |
Silvestrini M, Fruk L, Moretto LM, Ugo P. Detection of DNA hybridization by methylene blue electrochemistry at activated nanoelectrode ensembles. J Nanosci Nanotechnol. 2015;15(5):3437–3442. | |
Kamikawa TL, Mikolajczyk MG, Kennedy M, et al. Nanoparticle-based biosensor for the detection of emerging pandemic influenza strains. Biosens Bioelectron. 2010;26(4):1346–1352. | |
Silva MMS, Dias A, Cordeiro MT, Marques E, Goulart MOF, Dutra RF. A thiophene-modified screen printed electrode for detection of dengue virus NS1 protein. Talanta. 2014;128:505–510. | |
Chung D-J, Kim K-C, Choi S-H. Electrochemical DNA biosensor based on avidin-biotin conjugation for influenza virus (type A) detection. Appl Surf Sci. 2011;257(22):9390–9396. | |
Tam PD, Hieu NV, Chien ND, Le A-T, Tuan MA. DNA sensor development based on multi-wall carbon nanotubes for label-free influenza virus (type A) detection. J Immunol Meth. 2009;350(1–2):118–124. | |
Hughes MP, Morgan H, Rixon FJ, Burt JPH, Pethig R. Manipulation of herpes simplex virus type 1 by dielectrophoresis. Biochim Biophys Acta. 1998;1425(1):119–126. | |
Wang R, Lin J, Lassiter K, et al. Evaluation study of a portable impedance biosensor for detection of avian influenza virus. J Virol Meth. 2011;178(1–2):52–58. | |
Lin JH, Lum J, Wang RH, et al. A portable impedance biosensor instrument for rapid detection of avian influenza virus. Sensors. Piscataway, New Jersey: IEEE; 2010:1558–1563. | |
Chang H, Wang Y, Li J. Electrochemical DNA sensors: from nanoconstruction to biosensing. Curr Org Chem. 2011;15(4):506–517. | |
Lin CS, Chen YY, Cai ZX, Luo F, Wang YR, Chen X. An electrochemical biosensor for the sensitive detection of specific DNA based on a dual-enzyme assisted amplification. Electrochim Acta. 2014;147:785–790. | |
Xu MD, Zhuang JY, Chen X, Chen GA, Tang DP. A difunctional DNA-AuNP dendrimer coupling DNAzyme with intercalators for femtomolar detection of nucleic acids. Chem Commun. 2013;49(66):7304–7306. | |
Sahoo P, Suresh S, Dhara S, Saini G, Rangarajan S, Tyagi AK. Direct label free ultrasensitive impedimetric DNA biosensor using dendrimer functionalized GaN nanowires. Biosens Bioelectron. 2013;44:164–170. | |
Vlachova J, Tmejova K, Kopel P, et al. A 3D microfluidic chip for electrochemical detection of hydrolysed nucleic bases by a modified glassy carbon electrode. Sensors (Basel, Switzerland). 2015; 15(2):2438–2452. | |
Trefulka M, Ferreyra N, Ostatna V, Fojta M, Rivas G, Palecek E. Voltammetry of osmium end-labeled oligodeoxynucleotides at carbon, mercury, and gold electrodes. Electroanalysis. 2007;19(12):1334–1338. | |
Meng X, Xu M, Zhu J, Yin H, Ai S. Fabrication of DNA electrochemical biosensor based on gold nanoparticles, locked nucleic acid modified hairpin DNA and enzymatic signal amplification. Electrochim Acta. 2012;71:233–238. | |
Palecek E, Ostatna V, Pechan Z. Fifty five years of nucleic acid electrochemistry. Chem Listy. 2014;108(5):490–499. | |
Zari N, Amine A, Ennaji MM. Label-free DNA biosensor for electrochemical detection of short DNA sequences related to human papilloma virus. Anal Lett. 2009;42(3):519–535. | |
Pournaghi-Azar MH, Ahour F, Hejazi MS. Differential pulse voltammetric detection of hepatitis C virus 1a oligonucleotide chain by a label-free electrochemical DNA hybridization biosensor using consensus sequence of hepatitis C virus 1a probe on the pencil graphite electrode. Electroanalysis. 2009;21(16):1822–1828. | |
Huang K-J, Liu Y-J, Zhang J-Z. Aptamer-based electrochemical assay of 17 beta-estradiol using a glassy carbon electrode modified with copper sulfide nanosheets and gold nanoparticles, and applying enzyme-based signal amplification. Microchim Acta. 2015;182(1–2):409–417. | |
Yamanaka K, Saito M, Kondoh K, et al. Rapid detection for primary screening of influenza A virus: microfluidic RT-PCR chip and electrochemical DNA sensor. Analyst. 2011;136(10):2064–2068. | |
Peng H-P, Hu Y, Liu P, et al. Label-free electrochemical DNA biosensor for rapid detection of mutidrug resistance gene based on Au nanoparticles/toluidine blue-graphene oxide nanocomposites. Sens Actuator B Chem. 2015;207:269–276. | |
Wang J, Shi A, Fang X, Han X, Zhang Y. An ultrasensitive supersandwich electrochemical DNA biosensor based on gold nanoparticles decorated reduced graphene oxide. Anal Biochem. 2015;469:71–75. | |
Bi S, Cui Y, Dong Y, Zhang N. Target-induced self-assembly of DNA nanomachine on magnetic particle for multi-amplified biosensing of nucleic acid, protein, and cancer cell. Biosens Bioelectron. 2014;53:207–213. | |
Krejcova L, Hynek D, Kopel P, et al. Development of a magnetic electrochemical bar code array for point mutation detection in the H5N1 neuraminidase gene. Viruses Basel. 2013;5(7):1719–1739. | |
Qiu W, Xu H, Takalkar S, et al. Carbon nanotube-based lateral flow biosensor for sensitive and rapid detection of DNA sequence. Biosens Bioelectron. 2015;64:367–372. | |
Senapati S, Slouka Z, Shah SS, et al. An ion-exchange nanomembrane sensor for detection of nucleic acids using a surface charge inversion phenomenon. Biosens Bioelectron. 2014;60:92–100. | |
Gao AR, Zou NL, Dai PF, et al. Signal-to-noise ratio enhancement of silicon nanowires biosensor with rolling circle amplification. Nano Lett. 2013;13(9):4123–4130. | |
Shi L, Chu ZY, Dong XL, Jin WQ, Dempsey E. A highly oriented hybrid microarray modified electrode fabricated by a template-free method for ultrasensitive electrochemical DNA recognition. Nanoscale. 2013;5(21):10219–10225. | |
Rai V, Hapuarachchi HC, Ng LC, Soh SH, Leo YS, Toh CS. Ultrasensitive cDNA detection of dengue virus RNA using electrochemical nanoporous membrane-based biosensor. Plos One. 2012;7(8). | |
Ruan SP, Li ZJ, Qi HL, Gao Q, Zhang CX. Label-free supersandwich electrogenerated chemiluminescence biosensor for the determination of the HIV gene. Microchim Acta. 2014;181(11–12):1293–1300. | |
Jampasa S, Wonsawat W, Rodthongkum N, et al. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens Bioelectron. 2014;54:428–434. | |
Ahour F, Pournaghi-Azar MH, Alipour E, Hejazi MS. Detection and discrimination of recombinant plasmid encoding hepatitis C virus core/E1 gene based on PNA and double-stranded DNA hybridization. Biosens Bioelectron. 2013;45:287–291. | |
Liu G, Lao RJ, Xu L, et al. Single-nucleotide polymorphism genotyping using a novel multiplexed electrochemical biosensor with nonfouling surface. Biosens Bioelectron. 2013;42:516–521. | |
Qi XW, Gao HW, Zhang YY, Wang XZ, Chen Y, Sun W. Electrochemical DNA biosensor with chitosan-Co3O4 nanorod-graphene composite for the sensitive detection of staphylococcus aureus nuc gene sequence. Bioelectrochemistry. 2012;88:42–47. | |
Chen X, Hong CY, Lin YH, Chen JH, Chen GN, Yang HH. Enzyme-free and label-free ultrasensitive electrochemical detection of human immunodeficiency virus DNA in biological samples based on long-range self-assembled DNA nanostructures. Anal Chem. 2012;84(19):8277–8283. | |
Chebil S, Macauley N, Hianik T, Korri-Youssoufi H. Multiwalled carbon nanotubes modified by NTA-copper complex for label-free electrochemical immunosensor detection. Electroanalysis. 2013;25(3):636–643. | |
Rivas GA, Rubianes MD, Rodriguez MC, et al. Carbon nanotubes for electrochemical biosensing. Talanta. 2007;74(3):291–307. | |
Alipour E, Ghourchian H, Boutorabi SM. Gold nanoparticle based capacitive immunosensor for detection of hepatitis B surface antigen. Anal Meth. 2013;5(17):4448–4453. | |
Miodek A, Sauriat-Dorizon H, Chevalier C, Delmas B, Vidic J, Korri-Youssoufi H. Direct electrochemical detection of PB1-F2 protein of influenza A virus in infected cells. Biosens Bioelectron. 2014;59:6–13. | |
Miodek A, Vidic J, Sauriat-Dorizon H, et al. Electrochemical detection of the oligomerization of PB1-F2 influenza A virus protein in infected cells. Anal Chem. 2014;86(18):9098–9105. | |
Hong SA, Kwon J, Kim D, Yang S. A rapid, sensitive and selective electrochemical biosensor with concanavalin A for the preemptive detection of norovirus. Biosens Bioelectron. 2015;64:338–344. | |
Cavalcanti IT, Guedes MIF, Sotomayor M, Yamanaka H, Dutra RF. A label-free immunosensor based on recordable compact disk chip for early diagnostic of the dengue virus infection. Biochem Eng J. 2012;67:225–230. | |
Xie ZX, Huang JL, Luo SS, et al. Ultrasensitive electrochemical immunoassay for avian influenza subtype H5 using nanocomposite. Plos One. 2014;9(4):e94685. | |
Braustein HE, Braustein IE. Real time diagnostic point of care by amperometric immuno-biosensor kit by flow technology. Microfluidic Mems/Nems Sens Dev. 2014;58(40):1–17. | |
Huang CW, Huang YJ, Yen PW, et al. A CMOS wireless biomolecular sensing system-on-chip based on polysilicon nanowire technology. Lab Chip. 2013;13(22):4451–4459. | |
Luo YL, Nartker S, Miller H, et al. Surface functionalization of electrospun nanofibers for detecting E. coli O157:H7 and BVDV cells in a direct-charge transfer biosensor. Biosens Bioelectron. 2010;26(4):1612–1617. | |
Wang R, Wang Y, Lassiter K, et al. Interdigitated array microelectrode based impedance immunosensor for detection of avian influenza virus H5N1. Talanta. 2009;79(2):159–164. | |
Amano Y, Cheng Q. Detection of influenza virus: traditional approaches and development of biosensors. Anal Bioanal Chem. 2005; 381(1):156–164. | |
Jiang X, Spencer MG. Electrochemical impedance biosensor with electrode pixels for precise counting of CD4(+) cells: a microchip for quantitative diagnosis of HIV infection status of AIDS patients. Biosens Bioelectron. 2010;25(7):1622–1628. | |
Ren R, Leng CC, Zhang SS. A chronocoulometric DNA sensor based on screen-printed electrode doped with ionic liquid and polyaniline nanotubes. Biosens Bioelectron. 2010;25(9):2089–2094. | |
Fang XQ, Tan OK, Tse MS, Ooi EE. A label-free immunosensor for diagnosis of dengue infection with simple electrical measurements. Biosens Bioelectron. 2010;25(5):1137–1142. | |
Hnaien M, Diouani MF, Helali S, et al. Immobilization of specific antibody on functionalized gold electrode for rabies virus detection by electrochemical impedance spectroscopy. Biochem Eng J. 2008;39(3):443–449. | |
Hejazi MS, Pournaghi-Azar MH, Ahour F. Electrochemical detection of short sequences of hepatitis C 3a virus using a peptide nucleic acid-assembled gold electrode. Anal Biochem. 2010;399(1):118–124. | |
Malecka K, Grabowska I, Radecki J, et al. Electrochemical detection of avian influenza virus genotype using amino-ssDNA probe modified gold electrodes. Electroanalysis. 2013;25(8):1871–1878. | |
John SV, Rotherham LS, Khati M, Mamba BB, Arotiba OA. Towards HIV detection: novel poly(propylene imine) dendrimer-streptavidin platform for electrochemical DNA and gp120 aptamer biosensors. Int J Electrochem Sci. 2014;9(10):5425–5437. | |
Rickert J, Gopel W, Beck W, Jung G, Heiduschka P. A ‘mixed’ self-assembled monolayer for an impedimetric immunosensor. Biosens Bioelectron. 1996;11(8):757–768. | |
Berggren C, Bjarnason B, Johansson G. Capacitive biosensors. Electroanalysis. 2001;13(3):173–180. | |
Novakova Z, Orinakova R, Orinak A, Hvizdos P, Fedorkova AS. Elimination voltammetry as a new method for studying the SAM formation. Int J Electrochem Sci. 2014;9(7):3846–3863. | |
Paulo TDF, Abruna HD, Diogenes ICN. Thermodynamic, kinetic, surface pK(a), and structural aspects of self-assembled monolayers of thio compounds on gold. Langmuir. 2012;28(51):17825–17831. | |
Sartore L, Sassi A, Barbaglio M. Properties of carbon-black composite vapour detectors based on multifunctional polymers. J Balk Tribol Assoc. 2010;16(4):498–509. | |
Zeng H, Jiang Y, Xie G, Yu J. Polymer coated QCM sensor with modified electrode for the detection of DDVP. Sens Actuator B Chem. 2007;122(1):1–6. | |
Lin C-C, Chen L-C, Huang C-H, Ding S-J, Chang C-C, Chang H-C. Development of the multi-functionalized gold nanoparticles with electrochemical-based immunoassay for protein A detection. J Electroanal Chem. 2008;619:39–45. | |
Park J-Y, Park S-M. DNA hybridization sensors based on electrochemical impedance spectroscopy as a detection tool. Sensors. 2009; 9(12):9513–9532. | |
Mashhadizadeh MH, Talemi RP. A highly sensitive and selective hepatitis B DNA biosensor using gold nanoparticle electrodeposition on an Au electrode and mercaptobenzaldehyde. Anal Meth. 2014; 6(22):8956–8964. | |
Tung YT, Wu MF, Wang GJ, Hsieh SL. Nanostructured electrochemical biosensor for th0065 detection of the weak binding between the dengue virus and the CLEC5A receptor. Nanomed Nanotechnol Biol Med. 2014;10(6):1335–1341. | |
Mishra NN, Retterer S, Zieziulewicz TJ, et al. On-chip micro-biosensor for the detection of human CD4(+) cells based on AC impedance and optical analysis. Biosens Bioelectron. 2005;21(5):696–704. | |
Kiilerich-Pedersen K, Dapra J, Cherre S, Rozlosnik N. High sensitivity point-of-care device for direct virus diagnostics. Biosens Bioelectron. 2013;49:374–379. | |
Jarocka U, Sawicka R, Gora-Sochacka A, et al. An immunosensor based on antibody binding fragments attached to gold nanoparticles for the detection of peptides derived from avian influenza hemagglutinin H5. Sensors. 2014;14(9):15714–15728. | |
Wang RH, Wang Y, Lassiter K, et al. Interdigitated array microelectrode based impedance immunosensor for detection of avian influenza virus H5N1. Talanta. 2009;79(2):159–164. | |
Xu S. Electromechanical biosensors for pathogen detection. Microchim Acta. 2012;178(3–4):245–260. | |
Mecea VM. From quartz crystal microbalance to fundamental principles of mass measurements. Anal Lett. 2005;38(5):753–767. | |
Sauerbrey G. Use of a quartz vibrator for weighing thin layers on a microbalance. Z Phys. 1959;155(1959):206–222. | |
Vaughan RD, O’Sullivan CK, Guilbault GG. Sulfur based self-assembled monolayers (SAM’s) on piezoelectric crystals for immunosensor development. Fresenius J Anal Chem. 1999;364(1–2):54–57. | |
Kanazawa KK, Gordon JG. Frequency of a quartz microbalance in contact with liquid. Anal Chem. 1985;57(8):1770–1771. | |
Skladal P, Riccardi CD, Yamanaka H, da Costa PI. Piezoelectric biosensors for real-time monitoring of hybridization and detection of hepatitis C virus. J Virol Meth. 2004;117(2):145–151. | |
Li D, Wang J, Wang R, et al. A nanobeads amplified QCM immunosensor for the detection of avian influenza virus H5N1. Biosens Bioelectron. 2011;26(10):4146–4154. | |
Lee YG, Chang KS. Application of a flow type quartz crystal microbalance immunosensor for real time determination of cattle bovine ephemeral fever virus in liquid. Talanta. 2005;65(5):1335–1342. | |
Chu X, Zhao ZL, Shen GL, Yu RQ. Quartz crystal microbalance immunoassay with dendritic amplification using colloidal gold immunocomplex. Sens Actuator B Chem. 2006;114(2):696–704. | |
Kim YK, Lim SI, Choi S, Cho IS, Park EH, An DJ. A novel assay for detecting canine parvovirus using a quartz crystal microbalance biosensor. J Virol Methods. 2015;219:23–27. | |
Zhou XD, Liu LJ, Hu M, Wang LL, Hu JM. Detection of hepatitis B virus by piezoelectric biosensor. J Pharmaceut Biomed Anal. 2002;27(1–2):341–345. | |
Dell’Atti D, Zavaglia M, Tombelli S, et al. Development of combined DNA-based piezoelectric biosensors for the simultaneous detection and genotyping of high risk human papilloma virus strains. Clin Chim Acta. 2007;383(1–2):140–146. | |
Hong S-R, Jeong H-D, Hong S. QCM DNA biosensor for the diagnosis of a fish pathogenic virus VHSV. Talanta. 2010;82(3):899–903. | |
Yao CY, Wang YX, Fu WL. HBV DNA detection by RCA-QCM biosensor. Chem J Chin Univ. 2014;35(8):1635–1639. | |
Owen TW, Al-Kaysi RO, Bardeen CJ, Cheng Q. Microgravimetric immunosensor for direct detection of aerosolized influenza A virus particles. Sens Actuator B Chem. 2007;126(2):691–699. | |
Hewa TMP, Tannock GA, Mainwaring DE, Harrison S, Fecondo JV. The detection of influenza A and B viruses in clinical specimens using a quartz crystal microbalance. J Virol Meth. 2009;162(1–2):14–21. | |
Wangchareansak T, Sangma C, Ngernmeesri P, Thitithanyanont A, Lieberzeit PA. Self-assembled glucosamine monolayers as biomimetic receptors for detecting WGA lectin and influenza virus with a quartz crystal microbalance. Anal Bioanal Chem. 2013;405(20):6471–6478. | |
Lu CH, Zhang Y, Tang SF, et al. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens Bioelectron. 2012;31(1):439–444. | |
Chen L, Huang J, Lin C, Tai D, Wu T, inventors; TAI D (TAID-Individual), assignee. New method for the detection of dengue virus NS1 protein on gold electrode by using immunosensor. United States patent US TW200519371-A. June 16, 2005. | |
Liu YC, Wang CM, Hsiung KP, Huang CJ. Evaluation and application of conducting polymer entrapment on quartz crystal microbalance in flow injection immunoassay. Biosens Bioelectron. 2003;18(7):937–942. | |
Sin MLY, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev Mol Diagn. 2014;14(2):225–244. | |
Kobayashi M, Kikuchi N, Sato A. Optical tomography of fluorophores in dense scattering media based on ultrasound-enhanced chemiluminescence. Appl Phys Lett. 2015;106(2):21103–21103. | |
Tansi FL, Ruger R, Rabenhold M, Steiniger F, Fahr A, Hilger I. Fluorescence-quenching of a liposomal-encapsulated near- infrared fluorophore as a tool for in vivo optical imaging. J Vis Exp. 2015(95):e52136. | |
Mason C, Markusen JF, Town MA, Dunnill P, Wang RK. Doppler optical coherence tomography for measuring flow in engineered tissue. Biosens Bioelectron. 2004;20(3):414–423. | |
Obaton AF, Sanogo Y, Lautru J, Lievre M, Durocher JN, Dubard J. Development of a new optical reference technique in the field of biology. IEEE Trans Instrum Meas. 2013;62(4):837–844. | |
Ivers SN, Baranov SA, Sherlock T, et al. Depth-resolved imaging and detection of micro-retroreflectors within biological tissue using optical coherence tomography. Biomed Opt Express. 2010;1(2):367–377. | |
Trantum JR, Baglia ML, Eagleton ZE, Mernaugh RL, Haselton FR. Biosensor design based on Marangoni flow in an evaporating drop. Lab Chip. 2014;14(2):315–324. | |
Lee CK, Lin CW, Lin SM, et al. From an integrated biochip detection system to a defensive weapon against the SARS-CoV virus: OBMorph. In: Taylor DP, Liu J, McIlroy D, et al, editors. Nanoengineered Assemblies and Advanced Micro/Nanosystems. Vol 820. Warrendale, PA: Materials Research Society; 2004:249–260. | |
Pires NMM, Dong T, Hanke U, Hoivik N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors. 2014;14(8):15458–15479. | |
Kim MG, Shon Y, Lee J, et al. Double stranded aptamer-anchored reduced graphene oxide as target-specific nano detector. Biomaterials. 2014;35(9):2999–3004. | |
Padilla-Parra S, Matos PM, Kondo N, Marin M, Santos NC, Melikyan GB. Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proc Natl Acad Sci U S A. 2012;109(43):17627–17632. | |
Rossi AM, Wang L, Reipa V, Murphy TE. Porous silicon biosensor for detection of viruses. Biosens Bioelectron. 2007;23(5):741–745. | |
Grepstad JO, Kaspar P, Johansen IR, Solgaard O, Sudbo A. Detection of single nano-defects in photonic crystals between crossed polarizers. Opt Express. 2013;21(25):31375–31389. | |
Yan RJ, Lynn NS, Kingry LC, et al. Detection of virus-like nanoparticles via scattering using a chip-scale optical biosensor. Appl Phys Lett. 2012;101(16). | |
Heinze BC, Song JY, Lee CH, Najam A, Yoon JY. Microfluidic immunosensor for rapid and sensitive detection of bovine viral diarrhea virus. Sens Actuator B Chem. 2009;138(2):491–496. | |
Strong LH, Hall DB, Edson CM, Hiep-hoa N, Whitt MA, Varadi G. Application of field-modulated birefringence and light scattering to biosensing. In: Miller BL, Fauchet PM, editors. Frontiers in Biological Detection: From Nanosensors to Systems III. Vol 7888. Bellingham WA: SPIE; 2011. | |
Luo SC, Sivashanmugan K, Liao JD, Yao CK, Peng HC. Nanofabricated SERS-active substrates for single-molecule to virus detection in vitro: a review. Biosens Bioelectron. 2014;61:232–240. | |
Halas NJ, Lal S, Chang W-S, Link S, Nordlander P. Plasmons in strongly coupled metallic nanostructures. Chem Rev. 2011; 111(6):3913–3961. | |
Vendrell M, Maiti KK, Dhaliwal K, Chang Y-T. Surface-enhanced Raman scattering in cancer detection and imaging. Trends Biotechnol. 2013;31(4):249–257. | |
Lin YY, Liao JD, Ju YH, Chang CW, Shiau AL. Focused ion beam-fabricated Au micro/nanostructures used as a surface enhanced Raman scattering-active substrate for trace detection of molecules and influenza virus. Nanotechnology. 2011;22(18):185308. | |
Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006;6(11):2630–2636. | |
Lin YY, Liao JD, Yang ML, Wu CL. Target-size embracing dimension for sensitive detection of viruses with various sizes and influenza virus strains. Biosens Bioelectron. 2012;35(1):447–451. | |
Chang C-W, Liao J-D, Shiau A-L, Yao C-K. Non-labeled virus detection using inverted triangular Au nano-cavities arrayed as SERS-active substrate. Sens Actuator B Chem. 2011;156(1):471–478. | |
Kaminska A, Witkowska E, Winkler K, Dziecielewski I, Weyher JL, Waluk J. Detection of hepatitis B virus antigen from human blood: SERS immunoassay in a microfluidic system. Biosens Bioelectron. 2015;66:461–467. | |
Lim D-K, Jeon K-S, Hwang J-H, et al. Highly uniform and reproducible surface-enhanced Raman scattering from DNA-tailorable nanoparticles with 1-nm interior gap. Nat Nanotechnol. 2011;6(7):452–460. | |
Negri P, Chen G, Kage A, et al. Direct optical detection of viral nucleoprotein binding to an anti-influenza aptamer. Anal Chem. 2012;84(13):5501–5508. | |
Pang YF, Wang JF, Xiao R, Wang SQ. SERS molecular sentinel for the RNA genetic marker of PB1-F2 protein in highly pathogenic avian influenza (HPAI) virus. Biosens Bioelectron. 2014;61:460–465. | |
Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42(2):150–161. | |
Abbas A, Linman MJ, Cheng Q. New trends in instrumental design for surface plasmon resonance-based biosensors. Biosens Bioelectron. 2011;26(5):1815–1824. | |
Pattnaik P. Surface plasmon resonance – applications in understanding receptor-ligand interaction. Appl Biochem Biotechnol. 2005;126(2):79–92. | |
Englebienne P, Van Hoonacker A, Verhas M. Surface plasmon resonance: principles, methods and applications in biomedical sciences. J Spectros. 2003;17(2–3):255–273. | |
Liu Y, Wilson WD. Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. In: Fox KR, editor. Drug-DNA Interaction Protocols. 2nd ed. Vol 613. New York, NY: Humana Press; 2010:1–23. | |
Sakao Y, Nakamura F, Ueno N, Hara M. Hybridization of oligonucleotide by using DNA self-assembled monolayer. Colloids Surf B Biointerfaces. 2005;40(3–4):149–152. | |
Aqua T, Naaman R, Daube SS. Controlling the adsorption and reactivity of DNA on gold. Langmuir. 2003;19(25):10573–10580. | |
Brockman JM, Frutos AG, Corn RM. A multistep chemical modification procedure to create DNA arrays on gold surfaces for the study of protein-DNA interactions with surface plasmon resonance imaging. J Am Chem Soc. 1999;121(35):8044–8051. | |
Burgener M, Sanger M, Candrian U. Synthesis of a stable and specific surface plasmon resonance biosensor surface employing covalently immobilized peptide nucleic acids. Bioconjugate Chem. 2000;11(6):749–754. | |
Lee C-Y, Nguyen P-CT, Grainger DW, Gamble LJ, Castner DG. Structure and DNA hybridization properties of mixed nucleic acid/maleimide-ethylene glycol monolayers. Anal Chem. 2007;79(12):4390–4400. | |
Ermini ML, Scarano S, Bini R, et al. A rational approach in probe design for nucleic acid-based biosensing. Biosens Bioelectron. 2011;26(12):4785–4790. | |
Jin W, Lin X, Lv S, Zhang Y, Jin Q, Mu Y. A DNA sensor based on surface plasmon resonance for apoptosis-associated genes detection. Biosens Bioelectron. 2009;24(5):1266–1269. | |
Yao X, Li X, Toledo F, et al. Sub-attomole oligonucleotide and p53 cDNA determinations via a high-resolution surface plasmon resonance combined with oligonucleotide-capped gold nanoparticle signal amplification. Anal Biochem. 2006;354(2):220–228. | |
Sipova H, Zhang S, Dudley AM, Galas D, Wang K, Homola J. Surface plasmon resonance biosensor for rapid label-free detection of microribonucleic acid at subfemtomole level. Anal Chem. 2010;82(24):10110–10115. | |
Wang S, Yang H, Zhang H, et al. A surface plasmon resonance–based system to genotype human papillomavirus. Cancer Genet Cytogenet. 2010;200(2):100–105. | |
Kim SA, Kim SJ, Lee SH, et al. Detection of avian influenza-DNA hybridization using wavelength-scanning surface plasmon resonance biosensor. J Opt Soc Korea. 2009;13(3):392–397. | |
Teng J, Gu D, Xu Y, et al. Screening method for real-time detection of influenza-A virus in human throat swabs by surface plasmon resonance biosensor. In: Zhu R, editor. Medical Materials and Engineering. Vol 140. Dürnten, Switzerland: Trans Tech Publications; 2012:210–219. | |
Ritzefeld M, Sewald N. Real-time analysis of specific protein-DNA interactions with surface plasmon resonance. J Amino Acids. 2012; 2012:816032–816032. | |
Smith EA, Erickson MG, Ulijasz AT, Weisblum B, Corn RM. Surface plasmon resonance imaging of transcription factor proteins: interactions of bacterial response regulators with DNA arrays on gold films. Langmuir. 2003;19(5):1486–1492. | |
Tsoi PY, Yang MS. Surface plasmon resonance study of the molecular recognition between polymerase and DNA containing various mismatches and conformational changes of DNA-protein complexes. Biosens Bioelectron. 2004;19(10):1209–1218. | |
Wolf LK, Gao Y, Georgiadis RM. Kinetic discrimination of sequence-specific DNA – drug binding measured by surface plasmon resonance imaging and comparison to solution-phase measurements. J Am Chem Soc. 2007;129(34):10503–10511. | |
Fisher RJ, Rein A, Fivash M, et al. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72(3):1902–1909. | |
Baltzinger M, Sharma KK, Mely Y, Altschuh D. Dissecting the oligonucleotide binding properties of a disordered chaperone protein using surface plasmon resonance. Nucleic Acids Res. 2013;41(22):10414–10425. | |
Yang Y, Wang Q, Guo D. A novel strategy for analyzing RNA-protein interactions by surface plasmon resonance biosensor. Mol Biotechnol. 2008;40(1):87–93. | |
Xu J, Wan JY, Yang ST, et al. A surface plasmon resonance biosensor for direct detection of the rabies virus. Acta Vet BRNO. 2012;81(2):107–111. | |
Kumbhat S, Sharma K, Gehlot R, Solanki A, Joshi V. Surface plasmon resonance based immunosensor for serological diagnosis of dengue virus infection. J Pharmaceut Biomed Anal. 2010;52(2):255–259. | |
Zheng S, Kim D-K, Park TJ, Lee SJ, Lee SY. Label-free optical diagnosis of hepatitis B virus with genetically engineered fusion proteins. Talanta. 2010;82(2):803–809. | |
Vaisocherova H, Mrkvova K, Piliarik M, Jinoch P, Steinbachova M, Homola J. Surface plasmon resonance biosensor for direct detection of antibody against Epstein-Barr virus. Biosens Bioelectron. 2007;22(6):1020–1026. | |
Wang R, Zhao J, Jiang T, et al. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J Virol Meth. 2013;189(2):362–369. | |
Bai H, Wang R, Hargis B, Lu H, Li Y. A SPR aptasensor for detection of avian influenza virus H5N1. Sensors. 2012;12(9):12506–12518. | |
Mandenius C-F, Wang R, Aldén A, et al. Monitoring of influenza virus hemagglutinin in process samples using weak affinity ligands and surface plasmon resonance. Anal Chim Acta. 2008;623(1):66–75. | |
Suenaga E, Mizuno H, Penmetcha KKR. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens Bioelectron. 2012;32(1):195–201. | |
Faegh S, Jalili N, Sridhar S. A self-sensing piezoelectric microcantilever biosensor for detection of ultrasmall adsorbed masses: theory and experiments. Sensors. 2013;13(5):6089–6108. | |
Godin M, Tabard-Cossa V, Miyahara Y, et al. Cantilever-based sensing: the origin of surface stress and optimization strategies. Nanotechnology. 2010;21(7). | |
Hansen KM, Thundat T. Microcantilever biosensors. Methods. 2005;37(1):57–64. | |
Grogan C, Raiteri R, O’Connor GM, et al. Characterisation of an antibody coated microcantilever as a potential immuno-based biosensor. Biosens Bioelectron. 2002;17(3):201–207. | |
Fritz J, Baller MK, Lang HP, et al. Translating biomolecular recognition into nanomechanics. Science. 2000;288(5464):316–318. | |
Su M, Li SU, Dravid VP. Microcantilever resonance-based DNA detection with nanoparticle probes. Appl Phys Lett. 2003;82(20):3562–3564. | |
Bajwa N, Maldonado CJ, Thundat T, Passian A. Piezoresistive measurement of Swine H1N1 hemagglutinin peptide binding with microcantilever arrays. AIP Adv. 2014;4(3). | |
Xu D, Liu L, Guan J, et al. Label-free microcantilever-based immunosensors for highly sensitive determination of avian influenza virus H9. Microchim Acta. 2014;181(3–4):403–410. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.