Back to Journals » Infection and Drug Resistance » Volume 16
Mycobacterium tuberculosis Sub-Lineage 4.2.2/SIT149 as Dominant Drug-Resistant Clade in Northwest Ethiopia 2020–2022: In-silico Whole-Genome Sequence Analysis
Authors Mekonnen D , Munshea A, Nibret E , Adnew B, Getachew H, Kebede A, Gebrewahid A, Herrera-Leon S, Amor Aramendia A , Benito A, Abascal E, Jacqueline C, Aseffa A , Herrera-Leon L
Received 30 July 2023
Accepted for publication 9 October 2023
Published 26 October 2023 Volume 2023:16 Pages 6859—6870
DOI https://doi.org/10.2147/IDR.S429001
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Sandip Patil
Daniel Mekonnen,1,2 Abaineh Munshea,2,3 Endalkachew Nibret,2,3 Bethlehem Adnew,4 Hailu Getachew,5 Amiro Kebede,5 Ananya Gebrewahid,5 Silvia Herrera-Leon,6 Aranzazu Amor Aramendia,7 Agustín Benito,8 Estefanía Abascal,6 Camille Jacqueline,6,9 Abraham Aseffa,4 Laura Herrera-Leon6,10
1Department of Medical Laboratory Sciences, School of Health Science, College of Medicine and Health Sciences, Bahir Dar University, Bahir Dar, Ethiopia; 2Health Biotechnology Division, Institute of Biotechnology, Bahir Dar University, Bahir Dar, Ethiopia; 3Department of Biology, Bahir Dar University, Bahir Dar, Ethiopia; 4Armauer Hansen Research Institute, Addis Ababa, Ethiopia; 5Amhara Public Health Institute, Bahir Dar, Ethiopia; 6National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain; 7Fundación Mundo Sano, Madrid, Spain; 8National Center of Tropical Medicine, Institute of Health Carlos III, Centro de Investigación Biomédica En Red de Enfermedades Infecciosas, Madrid, Spain; 9European Public Health Microbiology Training Programme, European Centre for Disease Prevention and Control, Stockholm, Sweden; 10CIBER Epidemiologia y Salud Publica, Madrid, Spain
Correspondence: Daniel Mekonnen, Email [email protected]
Introduction: Drug resistance (DR) in Mycobacterium tuberculosis complex (MTBC) is mainly associated with certain lineages and varies across regions and countries. The Beijing genotype is the leading resistant lineage in Asia and western countries. M. tuberculosis (Mtb) (sub) lineages responsible for most drug resistance in Ethiopia are not well described. Hence, this study aimed to identify the leading drug resistance sub-lineages and characterize first-line anti-tuberculosis drug resistance-associated single nucleotide polymorphisms (SNPs).
Methods: A facility-based cross-sectional study was conducted in 2020– 2022 among new and presumptive multidrug resistant-TB (MDR-TB) cases in Northwest Ethiopia. Whole-genome sequencing (WGS) was performed on 161 isolates using Illumina NovaSeq 6000 technology. The SNP mutations associated with drug resistance were identified using MtbSeq and TB profiler Bioinformatics softwares.
Results: Of the 146 Mtb isolates that were successfully genotyped, 20 (13.7%) harbored one or more resistance-associated SNPs. L4.2.2.ETH was the leading drug-resistant sub-lineage, accounting for 10/20 (50%) of the resistant Mtb. MDR-TB isolates showed extensive mutations against first-line anti-TB drugs. Ser450Leu/(tcg/tTg) for Rifampicin (RIF), Ser315Thr/(agc/aCc) for Isoniazid (INH), Met306Ile/(atg/atA(C)) for Ethambutol (EMB), and Gly69Asp for Streptomycin (STR) were the leading resistance associated mutations which accounted for 56.5%, 89.5%, 47%, and 29.4%, respectively. The presence of both clustered and non-clustered drug resistance (DR) isolates indicated that the epidemics is driven by both new DR development and acquired resistance.
Conclusion: The high prevalence of drug-resistant TB due to geographically restricted sub-lineages (L4.2.2.ETH) indicates the ongoing local micro epidemics. The Mtb drug resistance surveillance system must be improved. Further evolutionary analysis of L4.2.2.ETH strain is highly desirable to understand evolutionary forces that leads L4.2.2.ETH in to high level DR and transmissible sub-lineage.
Keywords: Mycobacterium tuberculosis, L.4.2.2, SIT149, drug resistance, Ethiopia
Introduction
Antimicrobial resistance is a hidden global pandemic that shattered over 4.9 million people in 2019 alone, and the burden is highest, mainly in low-resource settings.1 Drug-resistant tuberculosis (DR-TB) caused by Mycobacterium tuberculosis (Mtb) complex (MTBC), which is resistant to one or more anti-TB drugs, is a leading global public health challenge.2 Multidrug resistant TB (MDR-TB) refers to TB patients infected with MTBC strains that is resistant to rifampicin (RIF) and isoniazid (INH).3 The MDR-TB mortality rate was estimated to be over 20%.4
Globally, an estimated 450000 incident cases of MDR-TB or rifampicin-resistant TB (MDR/RR-TB) have been reported by 2021, with a 3.1% increase since 2020.5 India (26%), the Russian Federation (8.5%) and Pakistan (7.9%) shared the highest global DR-TB case load.5 Based on a review of 24 articles in Ethiopia, the prevalence of ionized (INH) monoresistance and any INH resistance was gauged at 6.2% and 15.6%, respectively. Similarly, the RIF monoresistance and any RIF resistance was estimated at 2.3% and 9.7%, respectively. Further, the overall pooled prevalence of MDR TB was 10.8%.6
Unlike other prokaryotes in which drug resistance (DR) is mainly mediated by mobile genetic elements, Mtb DR is mediated by spontaneous mutations in the form of single nucleotide polymorphisms (SNP) in various regions of the bacterial genome.7,8 In addition, several intrinsic and extrinsic bacterial factors play a role in the evolution of DR. Some of these factors include delayed diagnosis, inadequate treatment, poor drug quality, patient noncompliance and pharmacodynamics, persistence, dormancy, cross-resistance, antagonistic epitasis, and biofilm formation.9 Surprisingly, DR-TB has mainly been reported in countries with well-functioning health systems and good treatment adherence policies. This suggests that drug resistance in MTBC is determined by the strain genetic background besides patient and programmatic related factors.8 The MTBC genotypes were geographically structured.10 Hence, the MTBC responsible for DR-TB varies across regions and countries.11–18
The Beijing genotype is the leading global DR-TB lineage19 with extensive widening in the Eurasia landmass.20 Different sub-lineages, including L4.2.2/SIT149, T3-ETH, Haarlem, CAS, EAI5, and L3/SIT25, have been identified in Ethiopia, showing different levels of resistance.21–24 It was noted that L4.2.2. ETH/SIT149 is known by different names in Ethiopian literatures. Some of these included Ethiopia_3,25 NW-ETH3,26 L4.2. ETH,27 SIT-149.28 Mtb isolates with sympatric host associations are more likely to be MDR than allopatric genotypes.29
While Africa is viewed as an evolutionary foci for the most recent common ancestor of MTBC, Asia is recognized as the evolutionary center for DR-Mtb. The World Health Organization (WHO) developed a comprehensive catalogue of MTBC mutations associated with first- and second-line anti-TB drugs using 38215 isolates with both phenotypic and genotypic data collected from 41 countries.30,31 Briefly, a mutation in rpoB, which encodes the β-subunit of RNA polymerase determines RIF resistance.31 Mutations in catalase-peroxidase (katG) and 2-trans-enoyl-acyl carrier protein reductase (inhA)-encoding genes are associated with INH resistance.32 While mutation at embCAB (for arabinosyl transferase) leads to Ethambutol (EMB) resistance,33 mutation at pncA which encodes for pyrazinamidase/nicotinamidase is associated with Pyrazinamide (PZA) resistance.34 Streptomycin (STR) resistance is determined by genes encoding ribosomal protein S12 (rpsL), 16S rRNA (rrs) and 7-methylguanosine (m7G) methyltransferase (gidB).35
The major lineages or sub-lineages responsible for most Mtb drug resistance in Ethiopia were not highlighted before. Furthermore, first-line anti-TB DR-related SNPs are poorly characterized. This research is an extension to our previously published article.36 While the published article36 focused on genomic diversity and transmission dynamics, the present article ascertained the leading drug resistance sub-lineages, explored and characterized mutations associated with drug resistance to first-line anti-TB drugs.
Materials and Methods
Study Design, Population, Period and Setting
This facility-based cross-sectional study was conducted using two groups of (newly diagnosed and presumptive MDR TB) isolates. The first group of isolates were obtained from new (with no previous history of anti-TB treatment) PTB and tuberculous lymphadenitis (TBLN) cases at Felege-Hiwot Comprehensive Specialized Hospital (FHCSH), Bahir Dar, Han and Shum-Abo Health Centers from February 2021 to June 2022. The second group of Mtb isolates obtained from presumptive MDR-TB patients (previously treated for TB, had failed anti-TB treatment, or had been in contact with MDR-TB patients), collected from June 2020 to June 2022, and stored in the Mycobacterium reference laboratory at the Amhara Public Health Institute (APHI).
Mycobacterium tuberculosis Culture and Colony Extraction
The stored isolates were re-grown using two MTBC growth media: Löwenstein-Jensen (LJ) medium37 and mycobacterium growth indicator tube (MGIT) BACTEC 960 detection system.38 When both LJ and MGIT-960 were positive with sufficient colonies, the colony was extracted from LJ; otherwise, MGIT-960 was used. From the day of positivity, the liquid culture was maintained at 37°C for an additional 7 days to enrich the MTBC population and make the MTBC cells aggregate and plainly visible in the transparent media. One milliliter of MGIT-960 medium containing MTBC aggregates was placed in 1.5 milliliter screw-cap vials. The MTBC colonies on LJ media were harvested using a sterile plastic loop and transferred to 1.5-mL screw-cap vials containing 1 mL of distilled water. Colonies were inactivated by heating at 95°C for 30 minutes.39 Heat-inactivated MTBC was transported to the National Center of Microbiology, Institute of Carlos III (ISCIII), Madrid, Spain for DNA extraction and whole-genome sequencing (WGS).
Mycobacterium tuberculosis DNA Extraction
The DNA extraction was carried out using manual NZY Tissue gDNA isolation kit (nzytech genes and enzymes, Lisboa, Portugal)40 and a robotic Maxwell RSC cultured Cell DNA extraction kit (Promega Biotech Ibérica S.L.).41 The following adjustments were made to the manufacturer’s instructions for DNA extraction using the NZY Tissue Kit. Heat-inactivated MTBC isolate was centrifuged for two minutes at 11000rpm. Following the removal of the supernatant, 180µL of pre-aliquoted lysis buffer and 60µL of reconstituted lysozyme (20 mg/mL) were added. The mixture was then vortexed and incubated at 37°C for 3h. After addition of 75ul of sodium dodecyl benzene sulfonate (SDS) 10x and 20ul µL proteinase K (20 mg/mL), the mixture was vortexed and incubated overnight at 56°C. The samples were subjected to the same pretreatment prior to DNA extraction using the robotic Maxwell RSC DNA kit-based extraction. After DNA extraction, a fluorometer (Promega Corporation) was used to determine the concentration of double-stranded DNA. NanoDrop spectrophotometer (Thermo Fisher Scientific) was used to determine purity at 260/280 absorbance ratio. A DNA quantity greater than 1ng/µL was used as the cutoff criterion for library preparation.
Library Preparation and Sequencing
The Nextera DNA Flex Library Prep Workflow was used to prepare the input DNA library, according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The Illumina NovaSeq 6000 was used for sequencing.42
Bioinformatics Analysis
The Bioinformatics Unit at the ISCIII performed quality control analysis using fastp; an ultra-fast all-in-one FASTQ preprocessor (QC, adapters, trimming, filtering, splitting); fastQC v.0.11.3 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic v.0.36 software.43 The reads were assembled using SPAdes v.3.8.044 and the quality of genome assembly was assessed using QUAST software.45
MTBseq 1.0.3 bioinformatics tool46 was used to map the genome against RefSeq (GCF_003287165.1_ASM328716v1). Briefly, mapping was performed using the Burrows-Wheeler Aligner-Maximum Exact Match (BWA-MEM) algorithm. Sorting, indexing, deleting probable PCR duplicates, and removing temporary files were accomplished using Sequence Alignment/Map tools (SAM tools). The Genome Analysis Toolkit (GATK) was used to recalibrate base calls and real readings around insertions or deletions. Subsequently, strain and group calling, variant calling/discovery, and parsing were conducted. Genome annotation was performed using Prokka.47
Variants are indicated using four reads mapped in each forward and reverse orientation, at 75% allele frequency, and at least four calls with a Phred score of at least 20. The final sample-specific module performs phylogenetic classification of the input sample(s) using Homolka et al and Coll et al SNP-based typing.48,49 In SNP-based barcoding, variant subsets are automatically generated and filtered for the repetitive regions and resistance-associated genes.46 Molecular drug resistance was determined using MtbSeq 1.0.346 Mykrobe predictor50 and a TB-profiler (Galaxy Version 4.1.1+ galaxy).
Multiple sequence alignment (MSA) was performed against H37Rv (GenBank accession number NC_000962.3) using a fast Fourier transform (MAFFT).51 SNP-based phylogenetic reconstruction was then performed using randomized accelerated maximum likelihood (RaxML) with the general time reversal (GTR) model and a bootstrap value of 1000.52 Ancient M. tuberculosis (Anc Mtb) was used as the root. The tree was edited using FigTree (https://github.com/rambaut/figtree/releases). The sub-lineages, resistance SNPs and associated drugs were summarized using tables and figures.
Results
Molecular Epidemiology of Drug Resistant M. tuberculosis
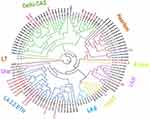
Of the 146 Mtb genotypes (66 from TBLN and 80 from PTB) successfully identified using the WGS technique,36 20 (13.7%) became resistant to at least one anti-TB drug. Of these, 18 were obtained from presumptive MDR-PTB cases and rest two were obtained from new PTB and TBLN cases (one from each). The proportion of MDR-TB was 17/146 (11.6%). Of the 20 resistant isolates, 10 (50%) belonged to L4.2.2. ETH, followed by Delhi-CAS (n = 6; 30%). The remaining four drug-resistant sub-lineages were L4.1.1.1/X-type, L4.3.4.1/LAM, L4.1.2.1/Haarlem, and L4.8/T (Figure 1). Further calculation of the proportion of drug resistance with respect to their sub-lineage confirmed that L4.2.2.ETH was still the leading drug-resistant isolate, 10/29 (34.5%). Similarly, the proportion of drug resistance in L3 was estimated at 14% (6/43). The phylogenetic tree depicts the distribution of DR Mtb sub-lineages (Figure 1).
Drug Resistance Profile of M. tuberculosis Sub-Lineages
Of the 20 isolates with one or more drug resistance mutations, 17 were MDR and three were RIF mono-resistant. No INH monoresistance and second-line anti-TB drug-resistance-associated mutations were identified. All ten isolates from L4.2.2. ETH demonstrated high-level MDR (MDR+: resistant to additional first-line drugs). A closer look at the drug resistance profile of each isolate showed that five L4.2.2. ETH/SIT149 sub-lineages were resistant to RIF (R), INH (H), ethambutol (EMB, E), pyrazinamide (PZA, Z) and streptomycin (STR, S); RHEZS. Two L4.2.2.ETH/SIT149 sub-lineages were resistant to RHEZ. Similarly, two L4.2.2.ETH sublineages harbored resistance mutation against RHES, and only one L4.2.2.ETH contained resistance determining mutation to RHE. Among the six lineage 3 resistant isolates, five were MDR (2 resistant for RHES, 2 resistant for RHE and one resistant for RH) and one was RIF monoresistance. All seven PZA-resistant isolates were from L4.2.2. ETH sub-lineages. In addition, the degree of resistance was low among generalist EA sub-lineages (X-type, Haarlem, LAM, and L4.8/T) compared to specialist sub-lineages (L4.2.2. ETH and some members of L3) in the present study (Table 1).
 |
Table 1 First-Line Anti-TB Drugs Resistant Sub-Lineages and Associated Amino Acid Substitutions /Mutation, Northwest Ethiopia, 2023 |
M. tuberculosis Drug Resistance Associated Mutations
Of the 24 RIF resistance SNPs, including the four double mutations, 54.2%, 12.5%, and 12.5% were associated with Ser450Leu (tcg/tTg), His445Arg (cac/cGc), and His445Tyr (cac/Tac) substitutions, respectively. Only katG mutation at codon 315 was associated with INH resistance. Of the 19 katG mutations, including two double SNPs at codon 315, Ser315Thr (agc/aCc) accounted for 89.5% (17/19), and the remaining 10.5% were for the Ser315Arg (agc/agA) substitution. Of the 20 drug-resistant sub-lineages, 15 developed EMB resistance via nine resistance conferring mutations. The dominant resistance-associated mutations in EMB were guanine with adenine (atg/atA) and guanine with cytosine (atg/atC) substitutions, both of which replaced methionine with isoleucine. Seven isolates from L4.2.2, were resistant to PZA in pncA gene. Mutations in two genes (rpsL and gidB) are responsible for STR resistance (Table 1 and Figure 2).
Collectively, out of 20 Mtb resistant isolates, 19 resistant isolates were identified from PTB, 11 (55%) were resistant to ≥4 first-line anti-TB drugs. L4.2.2.ETH were the leading DR clone. The resistance of L4.2.2.ETH for STR was mainly (8/9) mediated with low-level drug resistance conferring mutations, gibB (Table 1). Except STR resistance mutations, the prevalent mutations conferred high-level DR (Table 2).
 |
Table 2 Describe the Level of Resistance Associated with Each Resistance Marker, Northwest Ethiopia, 2023 |
Discussion
This study identified the leading drug-resistant sub-lineages and drug resistance associated SNPs. Furthermore, characterization of the mutations was performed. As such, this finding shows a relatively high proportion and degree of MDR in L4.2.2. ETH/SIT149/. Resistant isolates of L4.2.2/SIT149 have also been reported in Ethiopia,22–24 Saudi Arabia11 and Tanzania.17 The other prevalent DR Mtb lineage in Ethiopia is the Delhi-CAS (L3). Despite the widespread prevalence of DR L4.2.2/SIT149 isolates, much attention has not been paid to further characterization of downstream preventive and programmatic measures. It has been noted that L4.2.2/SIT149 and most Delhi-CAS isolates have sympatric relationships with Ethiopians and are more likely to be MDR than allopatric genotypes.29 The great southward human migration from Eurasia 3000 years before likely imported L4.2.2 into Ethiopia.27,58 Similarly, South Asia was predicted to be the origin of all L3 strains, with multiple independent introductions into North and Horn Africa.59 Taken together, the high prevalence of DR-TB due to Mtb sub-lineages with sympatric host associations indicates high rate of TB transmission taking place for long period of time.
Most of the MDR isolates from L4.2.2. ETH/SIT149 formed clusters. Unlike this report, Shea et al concluded that the clustering rate of DR-Mtb is relatively low compared to that of the susceptible strain because of the transmission fitness cost.53 As explained by Comas et al, fitness cost among dominant mutations is rare and MDR isolates are as competent as susceptible isolates.60 Hence, conclusions such as “MDR isolates are transmission deficient due to fitness cost” must be interpreted with caution.
In the present study, 11.6% of the patients had MDR-TB. According to a WHO report, the prevalence of MDR-TB in Ethiopia is 1.1% and 12% among new and previously treated cases, respectively.5 As such, Ethiopia was excluded from the WHO high-MDR-TB burden country list. However, the observation of high level of drug resistance among geographically restricted isolates is a warning sign that needs reassessment and further survey. Our WGS data showed that MDR isolates had extensive mutations, including double mutations. Such high-level mutations in multiple anti-TB drugs are proxy indicators of the possible occurrence of extensively drug-resistant TB in the near future.
Ser450Leu (tcg/tTg), Ser315Thr (agc/aCc), Met306Ile (atg/atA), and p.Gly69AspR were the dominant amino acids/SNPs substitutions responsible for resistance to RIF, INH, EMB, and STR, respectively. Based on the information in Table 2, except STR resistance determining mutation, gidB (p.Gly69AspR), all other prevalent mutations in this study confer high-level drug resistance. Dominant mutations have no fitness cost owing to compensatory mutations and confer high-level drug resistance.60 The characteristics and clinical interpretations of some of the prevalent mutations are presented below (Table 2).
High-level drug resistance is a complex evolutionary process that results in TB hotspot regions and inappropriate drug treatment settings.61 As described in Table 1 and Table 2, DR-Mtb strains develop high-level resistance to multiple first-line anti-TB drugs. Additionally, as shown in Figure 1, the DR strains contain both clustered and distinct isolates. This indicated that the epidemics of DR in the study area is driven by both independent evolution and transmission of acquired resistance. This again happens in countries where the implementation and adherence to WHO recommended TB control strategy is ineffective. Lower disease severity, low inflammation, high secondary case rate and high rate of MDR occurred among sympatric host pathogen association.29,62,63
The fitness of drug-resistant strains is heterogeneous.64 For instance, the fitness of DR Mtb strains showed comparable fitness with drug susceptible strains in some phylogeographic regions such as former Soviet Union.64 DR Mtb strains in this region might regain their pre-resistance fitness through compensatory evolution which again happen in areas where TB transmission takes place for long period of time.64
Limitation of the Study
Most of the drug-resistant sub-lineages were obtained from Mtb isolates archived in the laboratory. Hence, sampling bias is likely. Furthermore, the sample size was too small to obtain strong epidemiological information. Therefore, caution must be taken when interpreting the results.
Conclusion and Recommendations
Nineteen of the 20 drug-resistant isolates were obtained from PTB and L4.2.2. ETH/SIT149 was the dominant drug-resistant isolate followed by Delhi-CAS/L3. Furthermore, most of L4.2.2.ETH/SIT149 isolates were also resistant to all first-line anti-TB drugs, including STR. Proportionally, 56.5%, 89.5%, and 47% of RIF, INH, and EMB resistance-associated mutations were Ser450Leu/(tcg/tTg), Ser315Thr/(agc/aCc), and Met306Ile/(atg/atA(C)) substitutions, respectively. These mutations confer high levels of resistance. Mtb isolates that are resistant to all first-line anti-TB drugs, including STR, must be considered as a national threat. The high prevalence of drug resistance among geographically restricted sub-lineages indicates the ongoing local TB epidemics and uncontrolled TB transmission. Hence, active surveillance of DR-TB must be strengthened. Further evolutionary characterization of MDR isolates is very desirable to identify evolutionary forces driving the MDR-TB pandemic.
Data Sharing Statement
The WGS data are deposited in GenBank public database with bio project ID: PRJNA975069 and will be freely available shortly after publication of the manuscript.
Ethics Approval and Consent to Participate
This study was approved by the Research and Ethical Review Committee of Science College of Bahir Dar University (reference number PGRCSV/111/2012). Written informed consent was obtained from each participant prior to data collection. This study was conducted in accordance with the principles of the Declaration of Helsinki. All information obtained from the study participants was coded to maintain confidentiality.
Acknowledgments
The authors would like to express our gratitude to the Institute of Biotechnology and the Institute of Carlos III for their administrative support. Appreciation also goes to the International Federation for Clinical Chemistry (IFCC) for providing financial support to Daniel Mekonnen through the IFCC Professional Exchange Program (PEP). We also thank the Amhara Public Health Institute Mycobacteriology Laboratory staff, Felege Hiwote Referral Hospital DOTS clinic physicians, and the Pathology laboratory Team. DOTS clinic staff of Han Health, Bahir Dar, and Shum-Abo Health Centers also deserve special thanks for their support in the data collection. We also thank the Institute of Carlos III Mycobacteriology Laboratory staff, Martha and Rackel, for their support during the DNA extraction and WGS.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Sample collection was funded by the Institute of Biotechnology, Bahir Dar University, through the Endalkachew Nibret Mega Project. The Mtb culture and identification-related laboratory supply was supported by the Amhara Public Health Institute, Bahir Dar, Ethiopia. Whole-genome sequencing was performed with the great support of National Center of Microbiology, Institute of Carlos III, Madrid, Spain. The International Federation for Clinical Chemistry (IFCC) provided financial support to Daniel Mekonnen through the IFCC Professional Exchange Program (PEP) for three months stay in Madrid Spain for conducting the WGS analysis.
Disclosure
None of the authors has any relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending and royalties.
References
1. Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi:10.1016/S0140-6736(21)02724-0
2. World Health Organization. WHO Consolidated Guidelines on Tuberculosis: Module 4: Treatment -Drug-Resistant Tuberculosis Treatment 2022 Update. Geneva: World Health Organization; 2022.
3. Seung KJ, Keshavjee S, Rich ML. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb Perspect Med. 2015;5(9):a017863. doi:10.1101/cshperspect.a017863
4. Alemu A, Bitew ZW, Worku T, Gamtesa DF, Alebel A. Predictors of mortality in patients with drug-resistant tuberculosis: a systematic review and meta-analysis. PLoS One. 2021;16(6):e0253848. doi:10.1371/journal.pone.0253848
5. World Health Organization. Global Tuberculosis Report 2022. Geneva: World Health Organization; 2022.
6. Reta MA, Tamene BA, Abate BB, et al. Mycobacterium tuberculosis drug resistance in Ethiopia: an updated systematic review and meta-analysis. Trop Med Int Health. 2022;7(10):300. doi:10.3390/tropicalmed7100300
7. Almeida Da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. 2011;66(7):1417–1430. doi:10.1093/jac/dkr173
8. Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41(3):354–373. doi:10.1093/femsre/fux011
9. Müller B, Borrell S, Rose G, Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2013;29(3):160–169. doi:10.1016/j.tig.2012.11.005
10. Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(4):202–213. doi:10.1038/nrmicro.2018.8
11. Al-Ghafli H, Kohl TA, Merker M, et al. Drug-resistance profiling and transmission dynamics of multidrug-resistant Mycobacterium tuberculosis in Saudi Arabia revealed by whole genome sequencing. Infect Drug Resist. 2018;11:2219. doi:10.2147/IDR.S181124
12. Nguyen VAT, Bañuls A-L, Tran THT, et al. Mycobacterium tuberculosis lineages and anti-tuberculosis drug resistance in reference hospitals across Viet Nam. BMC Microbiol. 2016;16:1–9. doi:10.1186/s12866-016-0784-6
13. Zenteno-Cuevas R, Munro-Rojas D, Pérez-Martínez D, et al. Genetic diversity and drug susceptibility of Mycobacterium tuberculosis in a city with a high prevalence of drug resistant tuberculosis from Southeast of Mexico. BMC Infect Dis. 2021;21:1–12. doi:10.1186/s12879-021-06904-z
14. Bainomugisa A, Lavu E, Pandey S, et al. Evolution and spread of a highly drug resistant strain of Mycobacterium tuberculosis in Papua New Guinea. BMC Infect Dis. 2022;22(1):437. doi:10.1186/s12879-022-07414-2
15. Tan JL, Simbun A, Chan K-G, Ngeow YF. Genome sequence analysis of multidrug-resistant Mycobacterium tuberculosis from Malaysia. Scientific Data. 2020;7(1):135. doi:10.1038/s41597-020-0475-x
16. Ndungu PW, Kariuki S, Revathi G, Niemann S, Niemann S. Mycobacteria Interspersed repetitive units-variable number of tandem repeat, spoligotyping and drug resistance of isolates from pulmonary tuberculosis patients in Kenya. Advan Microbiol. 2017;7(3):205–216. doi:10.4236/aim.2017.73017
17. Kidenya BR, Mshana SE, Fitzgerald DW, Ocheretina O. Genotypic drug resistance using whole-genome sequencing of Mycobacterium tuberculosis clinical isolates from North-western Tanzania. Tuberculosis. 2018;109:97–101. doi:10.1016/j.tube.2018.02.004
18. Ssengooba W, Meehan CJ, Lukoye D, et al. Whole genome sequencing to complement tuberculosis drug resistance surveys in Uganda. Infect Genet Evol. 2016;40:8–16. doi:10.1016/j.meegid.2016.02.019
19. Keikha M, Majidzadeh M. Beijing genotype of Mycobacterium tuberculosis is associated with extensively drug-resistant tuberculosis: a global analysis. New Microbes and New Infections. 2021;43:100921. doi:10.1016/j.nmni.2021.100921
20. Merker M, Blin C, Mona S, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. doi:10.1038/ng.3195
21. Welekidan LN, Yimer SA, Skjerve E, et al. Whole genome sequencing of drug resistant and drug susceptible Mycobacterium tuberculosis isolates from Tigray region, Ethiopia. Front Microbiol. 2021;12:743198. doi:10.3389/fmicb.2021.743198
22. Worku G, Gumi B, Girma M, et al. Drug sensitivity of clinical isolates of Mycobacterium tuberculosis and its association with bacterial genotype in the Somali region, Eastern Ethiopia. Front Public Health. 2022;10:942618. doi:10.3389/fpubh.2022.942618
23. Diriba G, Kebede A, Tola HH, et al. Mycobacterial Lineages Associated with Drug Resistance in Patients with Extrapulmonary Tuberculosis in Addis Ababa, Ethiopia. Tuberc Res Treat. 2021;2021:1–7. doi:10.1155/2021/5239529
24. Ayalew S, Wegayehu T, Taye H, et al. Drug resistance conferring mutation and genetic diversity of Mycobacterium tuberculosis isolates in tuberculosis lymphadenitis Patients; Ethiopia. Infect Drug Resist;2021. 575–584. doi:10.2147/IDR.S298683
25. Tessema B, Beer J, Merker M, et al. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in Northwest Ethiopia: new phylogenetic lineages found in Northwest Ethiopia. BMC Infect Dis. 2013;13:131. doi:10.1186/1471-2334-13-131
26. Yimer SA, Namouchi A, Zegeye ED, et al. Deciphering the recent phylogenetic expansion of the originally deeply rooted Mycobacterium tuberculosis lineage 7. BMC Evol Biol. 2016;16(1):1–10. doi:10.1186/s12862-016-0715-z
27. Comas I, Hailu E, Kiros T, et al. Population genomics of Mycobacterium tuberculosis in Ethiopia contradicts the virgin soil hypothesis for human tuberculosis in Sub-Saharan Africa. Curr Bio. 2015;25(24):3260–3266. doi:10.1016/j.cub.2015.10.061
28. Firdessa R, Berg S, Hailu E, et al. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis, Ethiopia. Emerg Infect Dis. 2013;19(3):460. doi:10.3201/eid1903.120256
29. David S, Mateus A, Duarte EL, et al. Determinants of the sympatric host-pathogen relationship in tuberculosis. PLoS One. 2015;10(11):e0140625. doi:10.1371/journal.pone.0140625
30. Walker TM, Miotto P, Köser CU, et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe. 2022;3(4):e265–e73. doi:10.1016/S2666-5247(21)00301-3
31. World Health Organization. Catalogue of Mutations in Mycobacterium Tuberculosis Complex and Their Association with Drug Resistance. Geneva: World Health Organization; 2021.
32. Gurvitz A, Hiltunen JK, Kastaniotis AJ. Function of heterologous Mycobacterium tuberculosis InhA, a type 2 fatty acid synthase enzyme involved in extending C20 fatty acids to C60-to-C90 mycolic acids, during de novo lipoic acid synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74(16):5078–5085. doi:10.1128/AEM.00655-08
33. Perdigão J, Macedo R, Ribeiro A, Brum L, Portugal I. Genetic characterisation of the ethambutol resistance-determining region in Mycobacterium tuberculosis: prevalence and significance of embB306 mutations. Int J Antimicrob Agents. 2009;33(4):334–338. doi:10.1016/j.ijantimicag.2008.09.021
34. Juréen P, Werngren J, Toro J-C, Hoffner S. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2008;52(5):1852–1854. doi:10.1128/AAC.00110-08
35. Spies FS, Ribeiro AW, Ramos DF, et al. Streptomycin resistance and lineage-specific polymorphisms in Mycobacterium tuberculosis gidB gene. J Clin Microbiol. 2011;49(7):2625–2630. doi:10.1128/JCM.00168-11
36. Mekonnen D, Munshea A, Nibret E, et al. Comparative whole genome sequence analysis of mycobacterium tuberculosis isolated from pulmonary tuberculosis and tuberculous lymphadenitis patients in Northwest Ethiopia. Front Microbiol. 2023;14:1211267. doi:10.3389/fmicb.2023.1211267
37. Kent PT. Public Health Mycobacteriology: A Guide for the Level III Laboratory. US Department of Health and Human Services, Public Health Service, Centers; 1985.
38. Salman H, Siddiqi SR-G. MGIT TM Procedure Manual. USA and Germany: FIND Diagnostic; 2006.
39. Votintseva AA, Pankhurst LJ, Anson LW, et al. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53(4):1137–1143. doi:10.1128/JCM.03073-14
40. NZYtech. NZY Tissue gDNA Isolation kit Portuga; 2023. Available from: file:///C:/Users/Daniel%20m/Downloads/MB135_PB_V2101%20(1).pdf.
41. Corporation P. DNA Purification. Global; 2023.
42. Modi A, Vai S, Caramelli D, Lari M. The Illumina Sequencing Protocol and the NovaSeq 6000 System. Bacterial Pangenomics: Methods and Protocols. Springer; 2021:15–42.
43. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170
44. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
45. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi:10.1093/bioinformatics/btt086
46. Kohl TA, Utpatel C, Schleusener V, et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ. 2018;6:e5895. doi:10.7717/peerj.5895
47. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
48. Coll F, McNerney R, Guerra-Assunção JA, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5(1):1–5. doi:10.1038/ncomms5812
49. Homolka S, Projahn M, Feuerriegel S, et al. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One. 2012;7(7):e39855. doi:10.1371/journal.pone.0039855
50. Bradley P, Gordon NC, Walker TM, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun. 2015;6(1):1–15. doi:10.1038/ncomms10063
51. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi:10.1093/nar/gkf436
52. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi:10.1093/bioinformatics/btl446
53. Shea J, Halse TA, Kohlerschmidt D, et al. Low-level rifampin resistance and rpoB mutations in Mycobacterium tuberculosis: an analysis of whole-genome sequencing and drug susceptibility test data in New York. J Clin Microbiol. 2021;59(4):e01885–20. doi:10.1128/JCM.01885-20
54. Jagielski T, Bakuła Z, Brzostek A, et al. Characterization of mutations conferring resistance to rifampin in Mycobacterium tuberculosis clinical strains. Antimicrob Agents Chemother. 2018;62(10):e01093–18. doi:10.1128/AAC.01093-18
55. Napier G, Campino S, Phelan JE, Clark TG. Large-scale genomic analysis of Mycobacterium tuberculosis reveals extent of target and compensatory mutations linked to multi-drug resistant tuberculosis. Sci Rep. 2023;13(1):1–9. doi:10.1038/s41598-023-27516-4
56. Shen X, Shen G-M, Wu J, et al. Association between embB codon 306 mutations and drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51(7):2618–2620. doi:10.1128/AAC.01516-06
57. Wong SY, Lee JS, Kwak HK, et al. Mutations in gidB confer low-level streptomycin resistance in mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(6):2515–2522. doi:10.1128/AAC.01814-10
58. Mokrousov I. The quiet and controversial: ural family of Mycobacterium tuberculosis. Infect Genet Evol. 2012;12(4):619–629. doi:10.1016/j.meegid.2011.09.026
59. Shuaib YA, Utpatel C, Kohl TA, et al. Origin and global expansion of Mycobacterium tuberculosis Complex lineage 3. Genes. 2022;13(6):990. doi:10.3390/genes13060990
60. Comas I, Borrell S, Roetzer A, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44(1):106–110. doi:10.1038/ng.1038
61. Trauner A, Borrell S, Reither K, Gagneux S. Evolution of drug resistance in tuberculosis: recent progress and implications for diagnosis and therapy. Drugs. 2014;74(10):1063–1072. doi:10.1007/s40265-014-0248-y
62. Pasipanodya JG, Moonan PK, Vecino E, et al. Allopatric tuberculosis host–pathogen relationships are associated with greater pulmonary impairment. Infect Genet Evol. 2013;16:433–440. doi:10.1016/j.meegid.2013.02.015
63. Gagneux S, DeRiemer K, Van T, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103(8):2869–2873. doi:10.1073/pnas.0511240103
64. Borrell S, Gagneux S. Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect. 2011;17(6):815–820. doi:10.1111/j.1469-0691.2011.03556.x
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.