Back to Journals » Infection and Drug Resistance » Volume 16
Mycobacterium tuberculosis Rv0494 Protein Contributes to Mycobacterial Persistence
Authors Ji L , Jiang T, Zhao X, Cai D, Hua K, Du P, Chen Y, Xie J
Received 6 May 2023
Accepted for publication 14 July 2023
Published 22 July 2023 Volume 2023:16 Pages 4755—4762
DOI https://doi.org/10.2147/IDR.S419914
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Lei Ji,1 Tingting Jiang,1 Xin Zhao,2 Damin Cai,1 Kouzhen Hua,1 Peng Du,1 Yuanyuan Chen,3 Jianping Xie4
1School of Basic Medical Sciences and Forensic Medicine, Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China; 2Department of International Registration, Ustar Biotechnologies (Hangzhou) Ltd, Hangzhou, Zhejiang, People’s Republic of China; 3Tuberculosis Diagnosis and Treatment Center, Hangzhou Red Cross Hospital, Hangzhou, Zhejiang, People’s Republic of China; 4Institute of Modern Biopharmaceuticals, State Key Laboratory Breeding Base of Eco-Environment and Bio-Resource of the Three Gorges Area, Key Laboratory of Ministry of Education Eco-Environment of the Three Gorges Reservoir Region, Ministry of Education, Chongqing Municipal Key Laboratory of Karst Environment, School of Life Sciences, Southwest University, Chongqing, People’s Republic of China
Correspondence: Lei Ji, School of Basic Medical Sciences and Forensic Medicine, Hangzhou Medical College, No. 481 Binwen Road, Hangzhou, Zhejiang, 310053, People’s Republic of China, Tel +86 571 8769 2681, Fax +86 571 8769 2673, Email [email protected]
Purpose: Fatty acid metabolism plays an important role in the survival and pathogenesis of Mycobacterium tuberculosis. During dormancy, lipids are considered to be the main source of energy. A previous study found that Rv0494 is a starvation-inducible, lipid-responsive transcriptional regulator. However, the role of Rv0494 in bacterial persister survival has not been studied.
Methods: We constructed a Rv0494 deletion mutant strain of Mycobacterium tuberculosis H37Rv and evaluated the susceptibility of the mutant strain to antibiotics using a persistence test.
Results: We found that mutations in Rv0494 lead to survival defects of persisters, which reflected in increased sensitivity to isoniazid.
Conclusion: We conclude that Rv0494 is important for persister survival and may serve as a good target for developing new antibiotics that kill persister bacteria for improved treatment of persistent bacterial infections.
Keywords: drug target, drug tolerance, lipids, persisters
Introduction
Persisters are assumed to be defined as non-replicating or slow-growing bacteria that are not killed by antibiotics, which can return to a growth state upon removal of antibiotics, and are sensitive to the same antibiotics.1 Persisters play an important role in medicine pose a major challenge to the treatment of many bacterial infections such as tuberculosis and may be responsible for latent and persistent infections, long-term treatment and relapse after treatment.2 The mechanisms of persister formation are very complex and are not well understood.3
Bacteria have evolved various mechanisms to survive under stressful conditions. The success of Mycobacterium tuberculosis (M. tuberculosis, MTB) as a pathogen lies in its ability to survive for a long time in the host without the host itself showing any symptoms. It is to be reactivated only when the host immunity is compromised.4 During infection, MTB is exposed to harsh environmental conditions such as hypoxia, low pH and nutrient deprivation.5–7 Mycobacteria utilize their resources by efficiently coordinating gene expression according to prevailing conditions.8 The correct use of promoters in concert with transcriptional regulators plays an important role in mycobacterial physiology.9
FadR (Fatty acid metabolism regulator) acts as a sensor of the fatty acid level in bacteria. FadRs of Escherichia coli (E. coli), Vibrio vulnificus and Corynebacterium glutamicum have been extensively studied and are reported to play important roles in cell physiology and virulence.10–14 A FadR homologue in MTB, Rv0494, is a starvation-inducible, auto-regulatory FadR-like regulator.15 Fatty acid metabolism plays an important role in the survival and pathogenesis of MTB, as lipids are assumed to be the major source of energy during persistence.16 Rv0494 can negatively regulate the expression of kas operon genes, which are essential for mycolic acid biosynthesis in mycobacteria, it plays an inhibitory role in the regulation of fatty acid synthase II.17 It also functions as a self-regulator, which could bind to its own promoter regions.18 It was found that Rv0494, induced by starvation, is involved in regulating the expression of FabD and Rv2326c operons.15,17 Therefore, we assume that FadR plays a very important role in the persistence of MTB.
In this study, we constructed a Rv0494 deletion mutant (MUT) in MTB H37Rv, and exposed the mutants to various antibiotics to explore whether Rv0494 is important for persister survival. Our results demonstrate that Rv0494 is indeed involved in persister survival and tolerance to antibiotics.
Materials and Methods
Bacterial Strains and Growth Conditions
M. smegmatis mc2 155 was grown in Middlebrook 7H9 broth (BD) or on Middlebrook 7H10 agar (BD). MTB H37Rv was grown in Middlebrook 7H9 supplemented with OADC/ADC (Oadc oleic acid, albumin, dextrose and catalase medium/Albumin, dextrose and catalase medium) (BD) or on Middlebrook 7H10 supplemented with OADC. E. coli DH5α and E. coli HB101 were grown in LB broth (Solarbio) or on LB agar (Solarbio). When required, the following antibiotics were used at the specified concentrations: ampicillin 100 mg/L (Amresco), hygromycin (Hyg) 75 mg/L (Sigma), kanamycin 25 mg/L (MDBIO), isoniazid (INH) 4 mg/L (Merck) and rifampicin (RIF) 8 mg/L (Macklin). Ampicillin and kanamycin were used for the amplification of recombinant clones, plasmid isolation and transformation in E. coli, hygromycin was used for the screening of hygromycin-resistant transductants,19 isoniazid and rifampicin were used for antibiotic susceptibility testing. All oligonucleotides (Tsingke Biotechnology Co., Ltd.) and plasmids used in this study are listed in Table 1 and Table 2.
 |
Table 1 Primers Used in This Study |
 |
Table 2 Plasmids and Strains Used in This Study |
Construction of Rv0494 Knockout Strains and Complementation of Mutants
The kits used for gene knockout and complementation are listed in Table 3. All competent cells used in this research were prepared in house.19 The Rv0494 knockout mutant was constructed as described previously.19–21 Amplicons between 640 and 940 bases flanking the gene were PCR generated with primer sets (LFP/LRP and RFP/RRP (Table 1)) to generate gene-specific LHS (Left homology sequence) and RHS (Right homology sequence). Plasmid p0004s was digested with Van91I (Thermo Fisher), this fragment was ligated in one step to Van91I-digested LHS and RHS fragments corresponding to the upstream and downstream sequences flanking Rv0494. The ligation mix was transformed in E. coli DH5α, and the appropriate clones were confirmed by sequencing. One such clone was designated p0004s-AES (Allelic exchange substrates).
 |
Table 3 Kits Used in This Study |
After cleaving phAE159 and p0004s-AES, respectively, using the PacI (Thermo Fisher), the fragments were ligated. The ligation mix was transformed in E. coli HB101, single colonies growing on hygromycin-resistant plates were picked into LB+Hyg75 mg/L broth. The plasmids were extracted and identified using PacI restriction endonuclease digestion. Thus, the phAE159-AES phagemid was constructed.
The phagemid phAE159-AES was transformed into M. smegmatis mc2 155 to obtain phages that could be transfected into MTB H37Rv.19 Phages were transfected into MTB H37Rv and then screened for positive clones using Middlebrook 7H10+OADC+Hyg75mg/L. The primer sets (LYZFP/LYZRP and RYZFP/RYZRP (Table 1)) were then used to verify that the deletion mutants had been constructed.
Complementation of the Rv0494 knock-out mutants was performed utilizing the plasmid vector pMV361. A functional wild-type copy of Rv0494 was amplified by primers Rv0494FP and Rv0494RP. PCR products were digested with restriction enzymes EcoRI (Thermo Fisher) and HindIII (Thermo Fisher) and cloned into pMV361. The recombinant pMV361 containing Rv0494 (pMV361-Rv0494) was verified by DNA sequencing. The resulting constructs were transformed along with the empty vector pMV361 into mutant for complementation.
Antibiotic Susceptibility Testing
The susceptibilities of stationary phase Rv0494 mutants, complemented strains and the parent strain MTB H37Rv to various antibiotics, including INH (4 mg/L) and RIF (8 mg/L), were evaluated in drug exposure experiments in Middlebrook 7H9 supplemented with OADC. After incubating the bacterium in Middlebrook 7H9+OADC at 37°C without shaking for 3–4 weeks, the stationary phase cultures of Rv0494 mutants, complemented strains and the parent strain MTB H37Rv were obtained, and then INH and RIF were added to the cultures to give final concentrations of 4 mg/L and 8 mg/L, respectively. The cultures were continued to incubate at 37°C without shaking and CFU (Colony forming units) was determined after 0, 1, 3, 7 and 14 days of treatment. The number of CFU per milliliter was determined by plating serial dilutions of bacteria on Middlebrook 7H10+OADC plate. Three replications were performed for each set of data.
Results
Construction of Phagemid
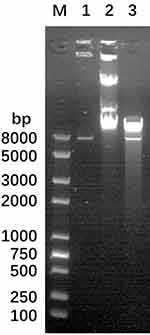
The LHS and RHS products were recovered by Van91I digestion and ligated with Van91I digested plasmid p0004s and transformed into E. coli DH5α. The resulting constructs p0004s-AES were screened and positive clones were sequenced. The positive clones obtained by identification were further cleaved by PacI and recovered, and ligated with the plasmid phAE159, also PacI cleaved, packaged and transformed into E. coli HB101 cells to screen for phagemid phAE159-AES, and the results of PacI cleavage validation are shown in Figure 1.
Phage-Based Mutant Construction
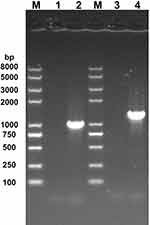
After transforming the phagemid phAE159-AES into M. smegmatis mc2 155, phages were obtained that could be transfected with MTB H37Rv. Finally, the ΔRv0494 strain was successfully completed using the phage we constructed. PCR was used to confirm the successful construction of the knockout strain (Figures 2 and 3). We performed amplification in wild-type and mutant strains using primers, respectively. The sequencing results proved that the mutants were successfully constructed.
Reduced Persister Levels of the Rv0494 Mutants in Antibiotic Exposure Assays
To determine the persister levels of the Rv0494 mutant, stationary phase cultures of the mutants (plus complement) and WT strain MTB H37Rv were exposed to no antibiotics as control (Figure 4a) and various antibiotics, including INH (4 mg/L) (Figure 4b) and RIF (8 mg/L) (Figure 4c), and the survival of the bacteria was monitored at different time points. At 14 days, hundred-fold reduction was noted for INH in the MUT. Overall, the results showed that the Rv0494 mutant was more susceptible than the wild-type strain MTB H37Rv to INH and that complementation of the Rv0494 mutant restored the level of persisters to close to wild-type levels in the antibiotic exposure assay (Figure 4). The RIF-treated mutant showed no significant persister reduction.
Discussion
Different phenotypes emerged after treatment of the Rv0494 mutant with different antibiotics. The mutant showed a significant persister reduction after INH treatment, suggesting that Rv0494 may play an important role in maintaining the persister in response to INH stress conditions. Both RIF and INH are the most important first-line anti-tuberculosis drugs; they are playing different roles in the treatment of tuberculosis. The prevailing view is that RIF binds to β subunit of DNA-dependent RNA polymerase and interferes with RNA transcription and elongation,22 INH is critically significant after stimulation acts to prevent mycolic acid synthesis and mycobacterial cell wall formation,23 they work in completely different ways. A recent study detected differentially expressed MTB proteins in RIF-related drug-resistant strains using label-free comparative proteomics approach.22 The gene data obtained in their experiments did not reveal any overlap with the genes regulated by Rv0494. Recently, a group used an evolutionary functional genomics approach identified lots of novel candidate regions involved in INH resistance in MTB.24 Among these candidate regions, they found that Rv1365c participates in INH resistance, this gene is also one of the genes regulated by Rv0494.17 This may explain why the same mutant strain was treated with RIF and INH at the same time, but showed different experimental results.
A recent research suggests that Rv0494 is a starvation-induced, auto-regulated FadR-like regulator, whereas this study was conducted after the strain entered the stationary phase with antibiotic stress applied, and it is expected that the absence of Rv0494 during this period resulted in the absence of its regulatory role, leading to the reduction of the persisters.15 We speculate that the deletion of Rv0494 in our mutant strain may have resulted in a partial loss of function of Rv1365c, thereby reducing MTB persistence. A recent study also revealed that Rv0586 of MTB is a true functional homologue of E. coli FadR, which complements E. coli ΔfadR.25 This result suggests to us that Rv0494 may play a more complex role in MTB, in a different manner from that of FadR in E. coli, and further studies are needed to address this hypothesis.
In another study for characterizing M. tuberculosis persisters, it was found that, when M. tuberculosis was treated with INH for a period of time, the expression of FabD was significantly upregulated in the wild-type strain.26 In a previous study, it was found that the expression of FabD was regulated by Rv0494,15 which plays a key role in the metabolism of type II fatty acid metabolism, and the absence of Rv0494, which compromises the function of FabD and thus affects fatty acid metabolism, may also be one of the reasons for the significant decrease in the number of colonies.
Conclusion
In this experiment, we just tested two of the most important anti-tuberculosis drugs, and subsequent testing of additional antibiotics (eg, pyrazinamide, ethambutol, streptomycin, etc.) is needed when studying their mechanism of action to clarify whether Rv0494 is a global regulator, which is related to persister formation. Since FadR is likely to be involved in persistence in other bacteria, our findings support the idea that Rv0494 protein could serve as a regulator contributing to mycobacterial persistence.
Acknowledgments
Thanks to the technical support from Shanghai Gene-optimal Science & Technology Co., Ltd.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 81701970, the Natural Science Foundation of Zhejiang Province, grant number LGF21H190005 and the China Scholarship Council.
Disclosure
Xin Zhao is employed by Ustar Biotechnologies (Hangzhou) Ltd. The authors report no other conflicts of interest in this work.
References
1. Hingley-Wilson SM, Ma N, Hu Y, et al. Loss of phenotypic inheritance associated with ydcI mutation leads to increased frequency of small, slow persisters in Escherichia coli. Proc Natl Acad Sci U S A. 2020;117(8):4152–4157.
2. Shi W, Zhang X, Jiang X, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–1632.
3. Li J, Ji L, Shi W, et al. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J Antimicrob Chemother. 2013;68(11):2477–2481.
4. Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12(8):581–591.
5. Papavinasasundaram KG, Anderson C, Brooks PC, et al. Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology. 2001;147(Pt 12):3271–3279.
6. Rohde K, Yates RM, Purdy GE, et al. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219:37–54.
7. Via LE, Lin PL, Ray SM, et al. Tuberculous granulomas are hypoxic in Guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76(6):2333–2340.
8. Martini MC, Zhou Y, Sun H, et al. Defining the transcriptional and post-transcriptional landscapes of Mycobacterium smegmatis in aerobic growth and hypoxia. Front Microbiol. 2019;10:591.
9. Rodrigue S, Provvedi R, Jacques PE, et al. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2006;30(6):926–941.
10. Agari Y, Agari K, Sakamoto K, et al. TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation. Microbiology. 2011;157(Pt 6):1589–1601.
11. Brown RN, Gulig PA. Regulation of fatty acid metabolism by FadR is essential for Vibrio vulnificus to cause infection of mice. J Bacteriol. 2008;190(23):7633–7644.
12. Casali N, White AM, Riley LW. Regulation of the Mycobacterium tuberculosis mce1 operon. J Bacteriol. 2006;188(2):441–449.
13. Georgi T, Engels V, Wendisch VF. Regulation of L-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J Bacteriol. 2008;190(3):963–971.
14. Santangelo MP, Blanco FC, Bianco MV, et al. Study of the role of Mce3R on the transcription of mce genes of Mycobacterium tuberculosis. BMC Microbiol. 2008;8:38.
15. Yousuf S, Angara R, Vindal V, et al. Rv0494 is a starvation-inducible, auto-regulatory FadR-like regulator from Mycobacterium tuberculosis. Microbiology. 2015;161(Pt 3):463–476.
16. Micklinghoff JC, Breitinger KJ, Schmidt M, et al. Role of the transcriptional regulator RamB (Rv0465c) in the control of the glyoxylate cycle in Mycobacterium tuberculosis. J Bacteriol. 2009;191(23):7260–7269.
17. Biswas RK, Dutta D, Tripathi A, et al. Identification and characterization of Rv0494: a fatty acid-responsive protein of the GntR/FadR family from Mycobacterium tuberculosis. Microbiology. 2013;159(Pt 5):913–923.
18. Minch KJ, Rustad TR, Peterson EJ, et al. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun. 2015;6:5829.
19. Bardarov S, Bardarov S, Pavelka MS, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148(Pt 10):3007–3017.
20. Jain P, Hsu T, Arai M, et al. Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. mBio. 2014;5(3):e01245–14.
21. Stover CK, de la Cruz VF, Fuerst TR, et al. New use of BCG for recombinant vaccines. Nature. 1991;351(6326):456–460.
22. Ullah N, Hao L, Banga Ndzouboukou JL, et al. Label-free comparative proteomics of differentially expressed Mycobacterium tuberculosis protein in rifampicin-related drug-resistant strains. Pathogens. 2021;10(5):54.
23. Jenkins HE, Zignol M, Cohen T. Quantifying the burden and trends of isoniazid resistant tuberculosis, 1994-2009. PLoS One. 2011;6(7):e22927.
24. Furio V, Moreno-Molina M, Chiner-Oms A, et al. An evolutionary functional genomics approach identifies novel candidate regions involved in isoniazid resistance in Mycobacterium tuberculosis. Commun Biol. 2021;4(1):1322.
25. Yousuf S, Angara RK, Roy A, et al. Mce2R/Rv0586 of Mycobacterium tuberculosis is the functional homologue of FadR(E. coli). Microbiology. 2018;164(9):1133–1145.
26. Jain P, Weinrick BC, Kalivoda EJ, et al. Dual-reporter Mycobacteriophages (Φ2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio. 2016;7(5):e01023–16.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.