Back to Journals » Infection and Drug Resistance » Volume 16
Mycobacterium colombiense Pneumonia in HIV-Infected Patients: Three Case Reports and a Literature Review
Authors Guo Y , Li X, Xiao Q, Yang J, Tao R, Xu L, Zhu B
Received 22 October 2023
Accepted for publication 19 December 2023
Published 22 December 2023 Volume 2023:16 Pages 7767—7773
DOI https://doi.org/10.2147/IDR.S441083
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yongzheng Guo,1 Xiaofeng Li,2 Qianggu Xiao,3 Jie Yang,4 Ran Tao,1 Lijun Xu,1 Biao Zhu1
1The Department of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, People’s Republic of China; 2The Department of Infectious Diseases, Huzhou Central Hospital, Huzhou, Zhejiang, People’s Republic of China; 3The Department of Infectious Diseases, Xiaoshan First People’s Hospital, Hangzhou, Zhejiang, People’s Republic of China; 4The Department of Infectious Diseases, Lishui Municipal Central Hospital, Lishui, Zhejiang, People’s Republic of China
Correspondence: Biao Zhu, Email [email protected]
Background: Mycobacterium colombiense pneumonia in HIV-infected patients is relatively unusual but is associated with a high mortality rate, as well as high rates of misdiagnosis and delayed diagnosis. Clinical metagenome next-generation sequencing (mNGS) may have potential for its accurate and timely diagnosis.
Case Presentation: We retrospectively reviewed the medical records of three HIV-infected patients who presented with M. colombiense pneumonia in Zhejiang Province between January 2019 and December 2020. No specific clinical presentations or radiological manifestations were found in any of the patients. The detection of M. colombiense is 28– 55 days earlier using mNGS on bronchoalveolar lavage fluid (BALF) compared to traditional culture methods. A combined treatment of rifabutin, clarithromycin, or azithromycin, and ethambutol did not provide timely relief of symptoms in these three patients. In the early stage of treatment, moxifloxacin and linezolid were used for several weeks. The average course of treatment for all three patients was close to 17 months.
Conclusion: We recommend early BALF mNGS for fast and accurate diagnosis of M. colombiense pneumonia in HIV-infected patients with low CD4 counts and long duration of symptoms. Further, moxifloxacin and linezolid may be beneficial in the early stage of treatment.
Keywords: metagenomic next-generation sequencing, nontuberculous mycobacteria, Mycobacterium colombiense, pneumonitis, HIV, AIDS
Introduction
Nontuberculous mycobacterium (NTM) infections can be life-threatening and are associated with a mortality rate of nearly 30% in HIV-infected patients, even in this era of highly active antiretroviral therapy.1,2 Pulmonary NTMs mainly affect HIV-infected patients with severe immunosuppression, and its delayed diagnosis is associated with poor prognosis.3–5 Thus, earlier diagnosis could help shorten the clinical course and improve survival outcomes. Currently, the diagnosis of NTM is mainly verified by polymerase chain reaction (PCR) and culture.6 However, the culture and PCR methods are time-consuming and have low sensitivity. Moreover, NTMs need to be identified at the species level, as different species usually require different therapeutic strategies. As a promising alternative, unbiased metagenomic next-generation sequencing (mNGS) offers a faster, culture-independent, and high-throughput methodology to detect potential pathogens from clinical specimens directly.7–11 mNGS has been used to identify uncommon or slow-growing pathogens that cause pneumonia.7–12 In our previous study, we found that the sensitivity of mNGS in the diagnosis of pneumonia in HIV-infected patients was significantly higher than that of conventional microbiological tests (79.6% vs 61.1%, P < 0.05).13 Thus, this technique could be useful for the accurate and timely diagnosis of NTM infections in HIV-infected patients.
Mycobacterium colombiense is a rare pathogen that infects patients with HIV infection, and it is associated with high rates of misdiagnosis, underdiagnosis, and delayed diagnosis. According to the existing case reports, the overall mortality rate of M. colombiense pulmonary infection or bacteraemia is significantly high.14–16 However, so far, reports on the identification of M. colombiense by mNGS in HIV-infected patients have been rare. The use of mNGS in the diagnosis of NTM in HIV-infected patients has been gradually increasing in our ward.13 Therefore, here we report three HIV-infected patients who had M. colombiense pneumonia and were diagnosed by mNGS.
Case Presentation
The demographic and clinical data of the three patients included in this report were obtained from the hospital electronic medical record system. Their written informed consent was obtained for the use of their data. Further, this study was approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine.
Case 1
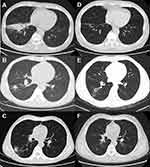
A 58-year-old man was hospitalized with a 4-month history of fever, expectoration, and weight loss of 7 kg. Three months before hospitalization, he had started antiretroviral therapy (ART) with a regime comprising dolutegravir (DTG), tenofovir (TDF), and lamivudine (3TC). His CD4 count was 9 cells/mm3. A pulmonary computed tomography (CT) scan showed interstitial lesions with infection, multiple nodules, and enlarged mediastinal lymph nodes (Figure 1A). The results of the serum T-SPOT, cryptococcal antigen, (1,3)-β-
Case 2
A 45-year-old HIV-infected man was hospitalized with a 3-month history of fever, expectoration, night sweats, and weight loss of 20 kg. His CD4 count was 112 cells/mm3. His pulmonary CT scan showed multiple nodular, flaky high-density shadows, and cavities in the lower lobe of the right lung (Figure 1B). The assay of serum, PBC, and BALF samples revealed negative results for common opportunistic microorganisms, such as Mycobacteria tuberculosis, NTM, Cryptococcus neoformans, Pneumocystis jirovecii, Candida, Aspergillus, Toxoplasma gondii, and Human Cytomegalovirus, among others. The results of BALF mNGS revealed the presence of M. colombiense based on 307 reads. The patient’s condition improved gradually after a 2-week combined regimen of Clr, EMB, Rfb, Mfx, and linezolid (LNZ). This was followed by an ART regime comprising DTG, TDF, and 3TC. Further, an oral Clr, EMB, Rfb, and Mfx regimen was prescribed after the patient was discharged. However, after initiation of ART for 18 days, the patient developed paradoxical NTM-immune reconstitution inflammatory syndrome (IRIS) and his CD4 count increased to 168 cells/mm3. CT-guided lung puncture biopsy was performed, and the sample was found to be positive for M. colombiense after 16 days of culture. LNZ was added to the anti-NTM therapy again, and the patient was discharged in good condition after a 12-day combined regimen of Clr, EMB, Rfb, Mfx, and LNZ. The anti-NTM treatment was continued for nearly 18 months, and his clinical and imaging results showed that the disease had resolved (Figure 1E).
Case 3
A 36-year-old young man was admitted to the hospital because he had high fever, dry cough, nausea, night sweats, and progressive fatigue over 3 weeks. He had started on ART with efavirenz (EFV), TDF, and 3TC 10 months ago. His pulmonary CT scan showed multiple flaky, nodular, high-density shadows in the right lung, along with enlarged mediastinal lymph nodes (Figure 1C). The routine screening of fungi, protozoa, and viruses through methods, such as blood culture, PCR, and serological tests, among others, revealed negative results. M. colombiense was identified based on 34 reads by mNGS of BALF. M. colombiense was identified from blood culture at 38 days after hospitalization. An anti-mycobacterial regimen comprising Mfx, AZI, Rfb, and EMB was prescribed, with emtricitabine/tenofovir (FTC/TDF) plus DTG for ART. The patient’s condition gradually improved, but he developed paradoxical NTM-IRIS approximately 1 month later. LNZ was then added to the anti-NTM therapy for about 2 weeks. The patient completed 14 months of treatment, after which the disease had resolved based on clinical and imaging observations (Figure 1F).
Discussion
Here, we have described three cases of M. colombiense pneumonia in HIV-infected patients at an advanced-stage. M. colombiense is classified as a slow-growing mycobacterium that is closely similar to the M. avium complex (MAC) based on its genetic sequence.16,17 Only a few reports of HIV-infected patients with M. colombiense infection have been published. The first cases were reported in three HIV-infected Colombian patients in 2006 (Table 1).14–16,18 No specific clinical and radiological manifestations have been identified in the reported cases so far, as observed in our cases too. In our series, two of the patients exhibited respiratory symptoms and various pulmonary lesions for three to four months before they were presented to the hospital, and one patient had been experiencing these symptoms for three weeks (Table 2). M. colombiense infection usually occurs in the advanced stage of HIV infection and is characterized by CD4 counts less than 50 cells/mm3.14,15,18 In our cases, the CD4 count of two patients (case 1 and case 3) was significantly lower than 50 cells/mm3, and the CD4 count in case 2 was slightly higher than 100 cells/mm3. All three patients were in the advanced stage of HIV infection, and their CD4 counts are consistent with those reported in the literature for advanced-stage HIV.14,15,18
 |
Table 1 Ten Previously Published Case Reports of HIV-Infected Patients with Mycobacterium colombiense Infection |
 |
Table 2 Demographic Data, Clinical Characteristics, and Outcome in Three Cases of HIV-Infected Patients with Mycobacterium colombiense Pneumonitis |
Early diagnosis of M. colombiense pneumonitis is a challenge because its symptoms are not specific and mimic those of a number of benign and malignant diseases.15,19 Moreover, due to the inhibitory effect of antibiotics used before specimen collection or the limited number of selected targets of molecular assays, the causative pathogen cannot be detected in time with the traditional detection methods. Furthermore, the limitations of conventional diagnostic tests may result in an underestimation of the prevalence of pulmonary M. colombiense. In our case series, mNGS allowed for a diagnosis of M. colombiense 28 to 55 days earlier than the traditional culture method. Thus, mNGS appears to have an advantage over traditional methods as it allows for early detection of pathogens and can, therefore, improve the prognosis of patients.
There is no consensus on the optimal anti-mycobacterial regimen for M. colombiense infection.16,20 Susceptibility tests have indicated that the drug susceptibility patterns of M. colombiense are similar to those of MAC, so therapeutic regimens for M. colombiense infections are mainly based on treatment regimens for MAC.14,16,20 In our patients, a combination of AZI or Clr, EMB, and Rfb was empirically prescribed as the initial therapy.21,22 The vitro susceptibility of Mfx and LNZ to MAC has been proven previously.23–25 Accordingly, the addition of Mfx and LNZ to the multidrug regimens were adopted in our study. However, as there are discrepancies between in vitro drug susceptibility test results and in vivo clinical outcomes; further studies are needed to evaluate the efficacy and the optimal course of Mfx and LNZ.6
Even though our case observations indicate the potential of mNGS, the results are limited because they were obtained from three different laboratories from different regions. Due to the lack of standard methods and availability of reference materials, the performance of different mNGS laboratories vary in terms of the identification of microbes and differentiation of true pathogens.26–28 At present, there is relatively little research in this area. Thus, further research is needed to improve the database setup and standardize the operation process.
Conclusions
In summary, we have reported three cases of HIV-infected adults with pulmonary M. colombiense infection who were diagnosed by mNGS and reviewed the associated literature. There were no specific clinical or radiological manifestations in these patients, but mNGS allowed for early diagnosis and identification of the pathogen. Thus, for patients in whom M. colombiense infection is suspected, BALF mNGS could allow for fast and accurate diagnosis of NTM. Further, although the patients responded to the administered anti-mycobacterial regime, the lung lesions regressed slowly and the infection recurred in two cases. Based on the observations in our cases, drug susceptibility testing is recommended for NTM infection patients if treatment regimens with Rfb, Clr, or AZI and EMB are ineffective. Alternatively, empirical addition of Mfx and LNZ may yield good results and could be considered.
Abbreviations
mNGS, metagenomic next-generation sequencing; NTM, Nontuberculous mycobacteria; M. colombiense, Mycobacterium colombiense; MAC, M. avium complex; BALF, bronchoalveolar lavage fluid; PBC, protected brush catheter; PCR, polymerase chain reaction; CT, computed tomography; ART, antiretroviral therapy; DTG, Dolutegravir; TDF, Tenofovir; 3TC, Lamivudine; EFV, efavirenz; FTC/TDF, emtricitabine/tenofovir; Mfx, moxifloxacin; AZI, azithromycin; Amk, Amikacin; Rfb, rifabutin; EMB, ethambutol; Clr, clarithromycin; LNZ, linezolid.
Data Sharing Statement
Patient data that were used in this study are provided in the tables.
Ethics Approval
The treatment of three patients was conducted in accordance with the Declaration of Helsinki. This report received the approval of the Ethics Committee of the First Affiliated Hospital of Zhejiang University School of Medicine (IIT20230802A).
Consent for Publication
Written informed consent has been obtained from three patients for the case details and images to be published. The case report does not contain any pictures that could identify the patient.
Acknowledgments
We especially acknowledge all frontline healthcare workers engaged in direct diagnosis, treatment, and care of patients.
Funding
This work was supported by the National Major Scientific and Technological Special Project for “Significant New Drugs Development” during the Thirteenth Five-Year Plan Period (grant number 2017ZX09303004-001). The funding organization had no involvement in the study or in the decision to submit the article for publication.
Disclosure
None of the authors have any conflicts of interest to declare.
References
1. Kobayashi T, Nishijima T, Teruya K, et al. High mortality of disseminated non-tuberculous mycobacterial infection in HIV-infected patients in the antiretroviral therapy era. PLoS One. 2016;11(3):e0151682. doi:10.1371/journal.pone.0151682
2. Bjerrum S, Oliver-Commey J, Kenu E, et al. Tuberculosis and non-tuberculous mycobacteria among HIV-infected individuals in Ghana. Trop Med Int Health. 2016;21(9):1181–1190. doi:10.1111/tmi.12749
3. World Health Organization. The top 10 causes of death. Available from: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
4. Griffiths EC, Pedersen AB, Fenton A, Petchey OL. The nature and consequences of coinfection in humans. J Infect. 2011;63(3):200–206. doi:10.1016/j.jinf.2011.06.005
5. Subahi EA, Aljafar MS, Barjas HH, Abdelrazek M, Rasoul FA. Co-infection by Cryptococcus neoformans fungaemia and non-tuberculous mycobacteria with Pneumocystis jiroveci pneumonia in a newly diagnosed HIV-infected patient. Clin Case Rep. 2021;9(7):e04191. doi:10.1099/jmm.0.000994
6. Panel on Opportunistic Infections in Adults and Adolescents with HIV. Guidelines for the prevention and treatment of opportunistic infections in adults and adolescents with HIV: recommendations from the centers for disease control and prevention. The National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. Available from: http://aidsinfo.nih.gov/contentfiles/lvguidelines/adult_oi.pdf.
7. Wang J, Han Y, Feng J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 2019;19(1):252. doi:10.1186/s12890-019-1022-4
8. Deurenberg RH, Bathoorn E, Chlebowicz MA, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. doi:10.1016/j.jbiotec.2016.12.022
9. Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. 2018;66(5):778–788. doi:10.1093/cid/cix881
10. Ramachandran PS, Cresswell FV, Meya DB, et al. Detection of Cryptococcus DNA by metagenomic next-generation sequencing in symptomatic cryptococcal antigenemia. Clin Infect Dis. 2019;68(11):1978–1979. doi:10.1093/cid/ciy1024
11. Xing XW, Zhang JT, Ma YB, Zheng N, Yang F, Yu SY. Apparent performance of metagenomic next-generation sequencing in the diagnosis of cryptococcal meningitis: a descriptive study. J Med Microbiol. 2019;68(8):1204–1210. doi:10.1099/jmm.0.000994
12. Zhou H, Larkin PMK, Zhao D, et al. Clinical impact of metagenomic next-generation sequencing of bronchoalveolar lavage in the diagnosis and management of pneumonia: a multicenter prospective observational study. J Mol Diagn. 2021;23(10):1259–1268. doi:10.1016/j.jmoldx.2021.06.007
13. Xie Y, Dai B, Zhou X, et al. Diagnostic value of metagenomic next-generation sequencing for multi-pathogenic pneumonia in HIV-infected patients. Infect Drug Resist. 2023;16:607–618. doi:10.2147/IDR.S394265
14. Lee MR, Chien JY, Huang YT, et al. Clinical features of patients with bacteraemia caused by Mycobacterium avium complex species and antimicrobial susceptibility of the isolates at a medical centre in Taiwan, 2008–2014. Int J Antimicrob Agents. 2017;50(1):35–40. doi:10.1016/j.ijantimicag.2017.02.016
15. Yu X, Jiang W. Mycobacterium colombiense and Mycobacterium avium complex causing severe pneumonia in a patient with HIV identified by a novel molecular-based method. Infect Drug Resist. 2021;14:11–16. doi:10.2147/IDR.S282190
16. Murcia MI, Tortoli E, Menendez MC, Palenque E, Garcia MJ. Mycobacterium colombiense sp. nov., a novel member of the Mycobacterium avium complex and description of MAC-X as a new ITS genetic variant. Int J Syst Evol Microbiol. 2006;56(Pt 9):2049–2054. doi:10.1099/ijs.0.64190-0
17. Gonzalez-Perez M, Murcia MI, Landsman D, Jordan IK, Marino-Ramirez L. Genome sequence of the Mycobacterium colombiense type strain, CECT 3035. J Bacteriol. 2011;193(20):5866–5867. doi:10.1128/JB.05928-11
18. Pena E, Machado D, Viveiros M, Jordao S. A case report of disseminated Mycobacterium colombiense infection in an HIV patient. Int J Mycobacteriol. 2019;8(3):295–297. doi:10.4103/ijmy.ijmy_100_19
19. Leguizamon J, Hernandez J, Murcia MI, Soto CY. Identification of potential biomarkers to distinguish Mycobacterium colombiense from other mycobacterial species. Mol Cell Probes. 2013;27(1):46–52. doi:10.1016/j.mcp.2012.08.009
20. Maurer FP, Pohle P, Kernbach M, et al. Differential drug susceptibility patterns of Mycobacterium chimaera and other members of the Mycobacterium avium-intracellulare complex. Clin Microbiol Infect. 2019;25(3):379 e1–379 e7. doi:10.1016/j.cmi.2018.06.010
21. Gordin FM, Sullam PM, Shafran SD, et al. A randomized, placebo-controlled study of rifabutin added to a regimen of clarithromycin and ethambutol for treatment of disseminated infection with Mycobacterium avium complex. Clin Infect Dis. 1999;28(5):1080–1085. doi:10.1086/514748
22. Griffith DE, Aksamit T, Brown-Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416. doi:10.1164/rccm.200604-571ST
23. Chen LC, Huang HN, Yu CJ, Chien JY, Hsueh PR. Clinical features and treatment outcomes of Mycobacterium chimaera lung disease and antimicrobial susceptibility of the mycobacterial isolates. J Infect. 2020;80(4):437–443. doi:10.1016/j.jinf.2020.01.005
24. Schulthess B, Schafle D, Kalin N, Widmer T, Sander P. Drug susceptibility distributions of Mycobacterium chimaera and other non-tuberculous mycobacteria. Antimicrob Agents Chemother. 2023;65(5). doi:10.1128/AAC.02131-20
25. Liu CF, Song YM, He WC, et al. Nontuberculous mycobacteria in China: incidence and antimicrobial resistance spectrum from a nationwide survey. Infect Dis Poverty. 2021;10(1):59. doi:10.1186/s40249-021-00844-1
26. Van Boheemen S, van Rijn AL, Pappas N, et al. Retrospective validation of a metagenomic sequencing protocol for combined detection of RNA and DNA viruses using respiratory samples from pediatric patients. J Mol Diagn. 2020;22(2):196–207. doi:10.1016/j.jmoldx.2019.10.007
27. Schlaberg R, Chiu CY, Miller S, et al. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141(6):776–786. doi:10.5858/arpa.2016-0539-RA
28. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi:10.1038/s41564-018-0349-6
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.