Back to Journals » International Journal of Nanomedicine » Volume 18
Multiple RNA Profiling Reveal Epigenetic Toxicity Effects of Oxidative Stress by Graphene Oxide Silver Nanoparticles in-vitro
Authors Yuan YG, Zhang YX, Liu SZ, Reza AMMT, Wang JL, Li L, Cai HQ, Zhong P, Kong IK
Received 20 October 2022
Accepted for publication 7 May 2023
Published 31 May 2023 Volume 2023:18 Pages 2855—2871
DOI https://doi.org/10.2147/IJN.S373161
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Farooq A. Shiekh
Yu-Guo Yuan,1,2,* Ya-Xin Zhang,1,* Song-Zi Liu,1,* Abu Musa Md Talimur Reza,3 Jia-Lin Wang,1,2 Ling Li,1,2 He-Qing Cai,1,2 Ping Zhong,1 Il-Keun Kong4
1College of Veterinary Medicine, Yangzhou University, Yangzhou, People’s Republic of China; 2Jiangsu Co-Innovation Center of Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, People’s Republic of China; 3Department of Molecular Biology and Genetics Faculty of Basic Sciences, Gebze Technical University, Kocaeli, Republic of Turkiye; 4Division of Applied Life Science (BK21 Four), Institute of Agriculture and Life Science, Gyeongsang National University, Jinju, Gyeongnam Province, Republic of Korea
*These authors contributed equally to this work
Correspondence: Yu-Guo Yuan; Il-Keun Kong, Email [email protected]; [email protected]
Introduction: The increasing industrial and biomedical utilization of graphene oxide silver nanoparticles (GO-AgNPs) raises the concern of nanosafety: exposure to the AgNPs or GO-AgNPs increases the generation of reactive oxygen species (ROS), causes DNA damage and alters the expression of whole transcriptome including mRNA, miRNA, tRNA, lncRNA, circRNA and others. Although the roles of different RNAs in epigenetic toxicity are being studied during the last decade, but still we have little knowledge about the role of circle RNAs (circRNAs) in epigenetic toxicity.
Methods: Rabbit fetal fibroblast cells (RFFCs) were treated with 0, 8, 16, 24, 32 and 48 μg/mL GO-AgNPs to test the cell viability and 24 μg/mL GO-AgNPs was selected as the experimental dose. After 24 h treatment with 24 μg/mL GO-AgNPs, the level of ROS, malondialdehyde (MDA), superoxide dismutase (SOD), intracellular ATP, glutathione peroxidase (GPx), and glutathione reductase (Gr) were measured in the RFFCs. High-throughput whole transcriptome sequencing was performed to compare the expression of circRNAs, long non-coding RNAs (lncRNA) and mRNA between 24 μg/mL GO-AgNPs-treated RFFCs and control cells. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed to validate the accuracy of circRNA sequencing data. Bioinformatics analyses were performed to reveal the potential functional roles and related pathways of differentially expressed circRNAs, lncRNA and mRNA and to construct a circRNA–miRNA–mRNA interaction network.
Results: We found that 57 circRNAs, 75 lncRNAs, and 444 mRNAs were upregulated while 35 circRNAs, 21 lncRNAs, and 186 mRNAs were downregulated. These differentially expressed genes are mainly involved in the transcriptional mis-regulation of cancer through several pathways: MAPK signaling pathway (circRNAs), non-homologous end-joining (lncRNAs), as well as PPAR and TGF-beta signaling pathways (mRNAs).
Conclusion: These data revealed the potential roles of circRNAs in the GO-AgNPs induced toxicity through oxidative damage, which would be the basis for further research to determine their roles in the regulation of different biological processes.
Keywords: circRNAs, GO-AgNPs, oxidative stress, epigenetic toxicity
Introduction
A wide variety of nanomaterials are either being used or are under investigation for their different industrial and biomedical uses as targeted drug delivery system, and antimicrobial agents against bacteria, fungi and viruses. For example, silver nanoparticles (AgNPs) are being used in different aspects of industrial and medical applications: such as food additives and food packaging, textiles and household appliances, cosmetics, medical devices, water disinfectants, and room sprays.1,2 The unique surface characteristics of AgNPs is the determining factor behind its antibacterial properties, which helps it to destroy the cell membranes or cell walls. However, application of AgNPs is limited because of its particle aggregation and cytotoxicity resulted in the host mammalian cells. Therefore, the combination of AgNPs and graphene-based nanocomposites such as reduced graphene oxide silver nanoparticles (GO-AgNPs) is being investigation as an alternative to the AgNPs.3,4 The hybrid GO-AgNPs could help us to avoid the AgNPs mediated cytotoxicity in mammalian cells.5 Now, the extensive application and production of these nanoparticles has increased our exposure to them via skin penetration, inhalation, or ingestion: thus, entering human cells either by endosomal uptake or by diffusion and inducing multiple unpredictable and deleterious health effects. Evidences indicate that AgNPs increase the generation of reactive oxygen species (ROS) and malondialdehyde (MDA), result in DNA damage, affect superoxide dismutase (SOD) and catalase (CAT) activity, induce differential expression of DNA replication, cellular senescence, and transcription of mitochondrial respiratory chain-related genes.6 Furthermore, surfactant‐coated AgNPs induces epigenetic changes which are associated with oxidative stress, MAPK signaling, and inflammation.7 Understanding the mechanism of how AgNPs induces epigenetic changes will help us to improve the design of AgNPs related nanoparticle, both in terms of harnessing their effects and minimizing the side-effects. Thus, conventional toxicity assays are not sufficient and integrated approaches are required to understand the complexities of cellular responses toward GO-AgNPs.
Number of studies showed the possible mechanisms of AgNPs mediated toxicity using both cells and animal models.5 AgNPs mediated cytotoxicity is possibly the result of one or a combination of the following effects on the mammalian cells: oxidative stress, inflammation response, DNA and molecular damage, growth inhibition, mitochondrial disruption, and changes in cell morphology.8 Although severity and dynamics of responses can vary among the cell types, but overall AgNPs causes a toxic response in varieties of cell types.9 However, the existing literature on genotoxicity of graphene family nanomaterials remains limited and conflicting. A few studies showed that graphene family nanomaterials had no adverse effects on genotoxicity.10,11 In contrast, many researchers have reported that the small size and sharp edges of graphene family nanomaterials can induce genotoxicity on culture cell.12,13 Compared to the well characterization of the cytotoxic effect of AgNPs, the epigenetic toxicity of AgNPs related nanoparticles on organisms remains poorly elucidated.14 Therefore, high-throughput analysis of whole transcriptome is necessary to understand the mechanism of GO-AgNPs inducing epigenetic toxicity.
Epigenetic changes could be understood by looking at the DNA methylation, histone tail modifications, and the activity of noncoding RNA, the latter includes different types of large and small non-coding RNAs that control the gene expression through varieties of mechanisms in response to internal or external stimuli.14 For instance, long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) are three different categories of ncRNA and are involved in the progression of different diseases.15 To date, AgNPs mediated epigenetic changes have been assessed to understand the nanosafety of AgNPs related nanoparticles exposure to culture cells and living animals.3,16–20 However, the changes of circRNAs and lncRNAs expression upon GO-AgNPs exposure to the cells and animals have not been investigated properly, and the nanosafety assessment of GO-AgNPs related nanomaterials is not yet completed.21
Knowing the epigenetic aspects of GO-AgNPs mediated toxicity will provide a better understanding of how nanomaterials impact genome integrity, and help us to develop practical strategies for the applications of nanomaterials in biomedicine and evaluations of nanomaterials mediated biotoxicity. We have already shown that 48 h treatment with GO-AgNPs incites DNA hypomethylation, increases the generation of ROS and induces apoptosis in caprine fetal fibroblast cells.22 In addition, long-term sublethal exposure of GO-AgNPs alters the expression of circRNAs and lncRNAs in caprine fetal fibroblast cells.23 However, we do not know how does the short-term exposure of GO-AgNPs influence the expression of circRNAs and lncRNAs. We expect to establish circRNAs and lncRNAs as biomarkers for GO-AgNPs mediated oxidative stress and toxicity in the cells. In this research, rabbit fetal fibroblast cells (RFFCs) were used as in vitro model to know the potential roles of GO-AgNPs mediated toxicity in cells and its impact on the expressions of circRNAs and lncRNAs.
Materials and Methods
Chemicals
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated. GO-AgNPs were synthesized and stored in our laboratory.22
The in-vitro Cell Culture Studies
RFFCs were isolated from 22-day-old fetuses that were recovered surgically from a New Zealand rabbit obtained from the Animal Genetic Engineering Laboratory of Yangzhou University as previously described.22 In brief, the head and internal organs of the fetus were removed and the remaining tissues of were dissociated into small pieces using scissors, washed three times with PBS, and cultured in Dulbecco’s Modified Eagle’s Medium/F12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin streptomycin (P/S) at 37°C in a humidified 5% CO2 incubator. For storage, the collected primary cells were frozen and stored in the liquid nitrogen: 3×106 cells/mL of DMEM/F-12 supplemented with 10% dimethyl sulfoxide (DMSO) and 20% FBS. Cells, between passages 2–4 were used for the experiments. The procedures of animal experiments were approved by the Animal Care and Use Committee of Yangzhou University, Yangzhou, China (license number: SYXK(Su)2017-0044 and protocol code 201803212).
Cell Viability Assay
The cell viability of GO-AgNPs treated RFFCs and its control counterpart was measured using the cell counting kit-8 (CCK-8; Rockville, MD, USA) according to the manufacturer’s instruction as described previously.22,23 In brief, the cell was seeded and cultured in 96-well plate in 100 μL complete medium containing different concentrations of GO-AgNPs (0, 8, 16, 24, 32 and 48 μg/mL) for 24 h with different concentrations (0, 8, 16, 24, 32 and 48 μg/mL) of GO-AgNPs. After that, 10 μL CCK-8 reagent was added to each well and incubated for 30 min at 37°C in dark. The absorbance was measured at 450 nm using a microplate reader (BioTek Synergy 2, USA). The reactions were conducted four times.
Determination of the Enzyme Activity
RFFCs were exposed to 24 μg/mL GO-AgNPs for 24 h and the cells were harvested to detect the activity of reactive oxygen species (ROS), malondialdehyde (MDA), superoxide dismutase (SOD), intracellular ATP, glutathione peroxidase (GPx), and glutathione reductase (Gr) as described in previous study.24–26 In brief, RFFCs were incubated in 10 μM DCFH-DA for 30 min at 37°C. After that, the cells were rinsed twice with PBS, suspended in 500 μL PBS and then the intracellular accumulation of ROS was measured using flow cytometry (Beckman-Coulter, USA). MDA was measured using a Lipid Peroxidation MDA Assay Kit. ATP levels were determined using the luciferin-luciferase-based ATP Assay Kit. GPx and Gr were determined using the Cellular Glutathione Peroxidase Assay Kit and Glutathione Reductase Assay Kit, respectively. All the assay kits were purchased from Beijing Solarbio Science & Technology, Beijing, China, and the assays were carried out according to the manufacturer’s instructions. The reactions were conducted four times.
Preparation of RNA Samples from RFFCs and Its Quality Assessment
Total RNA was isolated from three GO-AGNPs treated (24 μg/mL for 24 h) RFFC and three non-treated control samples using TRIzol reagent (Invitrogen, Carlsbad, CA, United States), according to the manufacturer’s protocol. The concentration and quality of the isolated total RNA were measured using NanoDrop ND-2000 spectrophotometer (NanaDrop, Wilmington, DE, United States). Furthermore, the integrity of the isolated RNA samples was assessed using standard denaturing agarose gel electrophoresis.
Generation of Illumina RNA-Seq Library and NovaSeq 6000 Sequencing
Construction of RNA-seq library as well as sequencing was performed by using the commercial service from CloudSeq Biotech Inc. (Shanghai, China). To maintain the integrity of RNA samples and deplete rRNAs, total RNAs were treated with RNase R (Epicentre, Madison, WI, USA) and the Ribo-Zero Magnetic Gold Kit (Epicentre, Madison, WI, USA), respectively. TruSeq Stranded Total RNA Library Prep Kit (Illumina, San Diego, CA, USA) was used to construct RNA-seq library from the rRNA-depleted total RNA samples according to the manufacturer’s protocols. The quality and quantity of the libraries were confirmed with the BioAnalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). After that, the libraries were denatured into single-stranded DNA molecules, captured on Illumina Flow Cells (Illumina) and then amplified in situ as clusters, and finally sequenced for 150 cycles using the NovaSeq 6000 Sequencing system (Illumina) as described previously.27
Bioinformatics Analysis
After the trimming of 30 adaptors and removal of low-quality reads, the high-quality reads were aligned to the reference genome with bowtie2 software, and circRNAs were detected and identified with find_circ software. For lncRNA and mRNA, high-quality reads were aligned to the rabbit reference genome (oryCun2) with hisat2 software (v2.0.4). HTSeq software (v0.9.1) was used to detect and identify lncRNA and mRNA. EdgeR software (v3.16.5) was used to normalize the data and perform analysis of differentially expressed circRNAs, lncRNA and mRNA. The raw junction reads were normalized to the number of total mapped reads and were log2 transformed. The differentially expressed circRNAs, lncRNA and mRNA (fold changes ≥ 2.0 and P < 0.05) were identified used for gene ontology (GO) and pathways analysis: Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics tool was applied for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis while Gene Ontology2 was applied to determine the biological roles of the differentially expressed circRNA, lncRNA and mRNA.23
Construction of circRNA–miRNA–mRNA Networks
The circRNA/miRNA interaction was predicted using Arraystar’s home-made miRNA target prediction software based on TargetScan and miRanda. The circRNA-miRNA network was constructed and visualized using Cytoscape v3.5.1. To explore the underlying mechanisms of top six differentially expressed circRNAs (Table 1 and Table 2), we predicted circRNA–miRNA–mRNA networks of five highest ranking miRNAs matched the potential miRNA binding sites of three down regulated and three upregulated circRNAs as described previously.23
 |
Table 1 Top 10 Down-Regulated circRNAs |
 |
Table 2 Top 10 Significantly Upregulated circRNAs |
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA samples were reverse-transcribed into cDNA with a random primer using SuperScriptTM III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. The expression of circRNAs was measured using quantitative polymerase chain reaction SYBR Green Master Mix (Takara, Dalian, China) in ABI 7500 thermocycler (Life Technologies, United States). Primers used for the qRT-PCR validation of the six differentially expressed circRNAs are enlisted in Table 3. Expression levels were calculated according to the 2−ΔΔCt method and GAPDH was used as reference gene. The amplification protocol is as follows: 5 min at 95°C followed by 39 cycles of 15 sec at 95°C and 1 min at 60°C. The reactions were performed in triplicate.
 |
Table 3 Primer Sequences for qRT-PCR Analysis |
Statistical Analysis
All of the experimental data are presented as mean ± standard deviation (SD), and the t-test was used for comparisons between two groups. p < 0.05 were considered as significantly different. GraphPad Prism5 software was used for statistical analysis.
Results
GO-AgNPs Induced Toxicity in RFFCs is Dependent on Dose
As shown in Figure 1, 24 h treatment of RFFCs with 8 µg/mL GO-AgNPs did not cause significant difference of its cell viability compared to the wildtype counterpart. However, increasing concentration of GO-AgNPs (16, 24, 32, and 48 µg/mL) significantly reduced (p < 0.05) the viability of RFFCS, indicating the dose-dependent effect of GO-AgNPs on the toxicity of RFFCs. For downstream experiments, 24 h treatment with 24 µg/mL GO-AgNPs was used based on IC50 value (50% inhibitory dose).
GO-AgNPs Alters the Metabolic and Enzymatic Activity in RFFCs
As shown in Figure 2A, generation of ROS significantly increased in RFFCs upon treatment with GO-AgNPs (p < 0.05). GO-AgNP treatment also increases the lipid peroxidation of the cell membrane (p < 0.05) compared to the control RFFCs (Figure 2B). These data indicate that GO-AgNPs induce oxidative stress to the cells. In addition, the intracellular ATP production significantly decreased (p < 0.05) in the GO-AgNPs treated RFFCs (Figure 2C), indicating that GO-AgNPs exposure potentially impairs the mitochondrial metabolism in RFFCs. Similarly, the activities of different metabolic enzymes including SOD (Figure 2D), GPx (Figure 2E) and Gr (Figure 2F) significantly decreased (p < 0.05) in the GO-AgNPs treated RFFCs. Overall, these results dictate that exposures to the GO-AgNPs alter activity of the metabolic enzymes that are involved in the detoxification of the cells.
GO-AgNPs Induces Differential Expression of mRNAs, lncRNAs and circRNAs in RFFCs
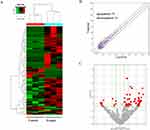
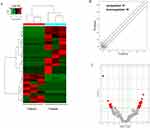
As shown in Figures 3–5, the expression patterns of hierarchical cluster analysis and differential expressions of mRNAs (Figure 3A–C), lncRNAs (Figure 4A–C) and circRNAs (Figure 5A–C) are detected between GO-AgNPs treated RFFCs and their control counterparts. Further details of the differentially expressed circRNAs are provided in Table 1 and Table 2. In brief, out of 630 differentially expressed mRNAs 444 mRNA upregulated and 186 mRNA downregulated, while fewer numbers of lncRNAs (75 upregulated and 21 downregulated) and circRNAs (57 upregulated and 35 downregulated) showed differential expression. To focus on the most important circRNAs, the top 10 downregulated and upregulated circRNAs in GO-AgNPs treated RFFCs are short-listed in Table 1 and Table 2, respectively. Out of these twenty differentially expressed circRNAs, 3 are intronic while 17 are sense overlapping.
Differentially Expressed mRNAs, lncRNAs and circRNAs are Important Parts of Cellular, Biological, and Metabolic Signalings
As shown in Figures 6–8, the differentially expressed genes are part of various biological processes (BP), cellular components (CC), and metabolic functions (MF), they are also playing regulatory roles in different signaling pathways. The down- and up-regulated circRNAs showed highest enrichment scores for intracellular protein transport and macromolecule localization (BP), nucleus and cytosol (CC), and ribosome binding and protein binding (MF). The differentially expressed lncRNA showed highest enrichment for double-strand break repair via nonhomologous end joining (BP), organelle membrane contact site (CC), and protein binding (MF). However, the differentially expressed mRNA molecules are mainly involved in the cellular protein metabolic process and microtubule nucleation (BP), catalytic complex and rough endoplasmic reticulum (CC), beta-tubulin binding and oxidoreductase activity, acting on paired donors with incorporation or reduction of molecular oxygen (MF). On the other hand, the KEGG pathway analysis showed that the differentially expressed circRNAs, lncRNAs and mRNAs are involved in the transcriptional misregulation in cancer and MAPK signaling pathway, non-homologous end-joining pathway, and PPAR and TGF-beta signaling pathways, respectively. It also dictates their potential involvement in the regulation of endometrial cancer as well as in the maintenance of pluripotency of stem cells.
Figure 6 Continued.
 |
Figure 8 Continued. |
Differentially Expressed circRNAs Potentially Regulates FoxO Signaling, MAPK Signaling, and Oxidative Phosphorylation
To understand the potential mechanism how differentially expressed circRNAs interacts with the miRNAs and mRNAs, sequences of six differentially expressed circRNAs were analyzed using TargetScan and an interacting network of circRNA–miRNA–mRNA was generated. To keep the constructed map comprehensible, it only illustrated the networks of the highest-ranking miRNAs that play regulatory roles on the differentially expressed mRNAs (Figure 9). It could be stipulated from the constructed network that the differentially expressed circRNAs potentially regulate the epigenetic toxicity through FoxO signaling pathway, MAPK signaling pathway, and oxidative phosphorylation.
Validation by qRT-PCR Proves the Accuracy of the RNA-Seq Data
Six differentially expressed circRNAs were selected for the validation by qRT-PCR: three downregulated circRNA genes (chrUn0019:1784368-1787145+, chr2:62054752-62070598- and chrUn0145:595264-602252+) and three upregulated circRNA genes (chrUn0017:2492594-2500768+, chrUn0029:1202788-1217961+, and chr13:133595 155-133605230+). As shown in Figure 10, the qRT-PCR expression level of the selected six circRNAs is consistent with the RNA-seq expression level of those circRNAs. This means that the RNA-seq results are accurate and reliable.
Discussion
This research clearly showed that GO-AgNPs may cause epigenetic toxicity by altering the expression of non-coding RNAs including circRNAs and lncRNAs. Oxidative stress mediated toxicity in cells is usually measured by the activity of the endogenous antioxidative factors such as CAT, SOD, GPx, and sulfhydryl form glutathione (GSH).25,26 Here, we have shown that GO-AgNPs increase oxidative stress in RFFCs by altering its metabolic and enzymatic activity. In particular, the generation of ROS increases which may impair cellular components such as DNA damage, activation of antioxidant enzymes, depletion of antioxidant molecules.28,29 The data showed that higher GO-AgNPs concentration increases the level of toxicity, this is potentially because of the GO-AgNPs mediated differential regulation of antioxidant activity.30–32 This means higher doses of GO-AgNPs increase the numbers of oxygen radicals which increases the susceptibility of RFFCs to cytotoxicity. We have also found that GO-AgNPs reduce ATP content and metabolic enzymes which is a clear sign of compromised mitochondrial functions.2,25 Since ATP is required for a cascade of events requiring phosphorylation of several proteins taking part in repair of DNA damage,33 it is logical to think that GO-AgNPs mediated depletion of ATP may compromise the DNA repair process in RFFCs.
Now, the question is whether the GO-AgNPs mediated toxicity is regulated at the epigenetic level. Previous studies related to the AgNPs mediated epigenetic toxicity mainly focuses on DNA and histone modifications, or alterations of miRNA expression.34–37 In this unique research, we have shown that GO-AgNPs cause differential expression of circRNAs and lncRNAs in RFFCs. In addition, we have constructed a comprehensive network map of circRNA–miRNA–mRNA that shows the potential scenario of how circRNAs are regulating gene expression by interacting with the miRNAs. Previous studies also showed that AgNPs related DNA damage could be measured by looking at the expression of related genes,38–40 lncRNA network,41,42 and circRNAs network.43 Our analysis (GO and KEGG pathways) indicates that the differentially expressed circRNA–miRNA–mRNA network is connected to the DNA damage and metabolism-related pathways. In particular, non-homologous end-joining, apoptosis, FoxO signaling, MAPK signaling, and oxidative phosphorylation are important pathways that are connected to the top differentially expressed circRNAs. MAPK signaling triggers cell proliferation, pro-inflammatory cytokines, oxidative stress and apoptosis in case of AgNPs induced toxicity.44–46 This means that the differential expression of circRNAs clearly reflects the cytotoxicity of the GO-AgNPs treated RFFCs. Therefore, we believe that the differentially expressed mRNA, lncRNAs, and circRNAs are associated with GO-AgNPs induced toxicity in cells.
In addition, the constructed circRNA–miRNA–mRNA networks could be connected to certain cellular, biological, and metabolic activity of the cells, including nucleobase-containing compound metabolic process, GTP metabolic process, and protein transports. For example, upregulation of evamiR-N6, eva-miR-N7 and eva-miR-N8 potentially depresses the ATP synthesis,25 which is dependent on the integrity of the mitochondrial membrane.47 KEGG pathway analysis also connected several differentially expressed circRNAs to the transcriptional misregulation which is related to the progression of cancers.25,48 Thus, the differentially expressed circRNAs in the GO-AgNPs treated RFFCs potentially interact with different miRNAs which regulate the metabolic activity and cytotoxicity of cells through multiple pathways.
Conclusions
In conclusion, GO-AgNPs increased the level of ROS, which induces cytotoxicity in RFFCs through lipid peroxidation and mitochondrial dysfunction. However, this cytotoxicity is potentially regulated at the epigenetic level through a complex interacting network of circRNA–miRNA–mRNA. This is a novel insight that will trigger further in vitro and in vivo studies to reveal the mechanisms of GO-AgNPs mediated epigenetic toxicity in cells. In particular, the role of circRNAs in the regulation of MAPK signaling is required to be investigated further.
Abbreviations
AgNPs, silver nanoparticles; circRNAs, circular RNAs; GO-AgNPs, reduced graphene oxide silver nanoparticles; GO, Gene Ontology; GPx, glutathione peroxidase; Gr, glutathione reductase; KEGG, Encyclopedia of Genes and Genomes; lncRNAs, long non-coding RNAs; MDA, malondialdehyde; miRNAs, microRNAs; Quantitative real-time polymerase chain reaction (qRT-PCR); RFFCs, rabbit fetal fibroblast cells; ROS, reactive oxygen species; SOD, superoxide dismutase.
Funding
This project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Development of a new precise cytosine base editor (JBGS〔2021〕025), the 111 Project D18007, Yangzhou city and Yangzhou University corporation (YZ2021161/2022187), and by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (grant no. RS-2023-00208894 and RDA no.RS-2023-00237137).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Prasath S, Palaniappan K. Is using nanosilver mattresses/pillows safe? A review of potential health implications of silver nanoparticles on human health. Environ Geochem Health. 2019;41(5):2295–2313. doi:10.1007/s10653-019-00240-7
2. Yuan YG, Peng QL, Gurunathan S. Effects of silver nanoparticles on multiple drug-resistant Strains of staphylococcus aureus and pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int J Mol Sci. 2017;18(3):569. doi:10.3390/ijms18030569
3. Wu K, Zhou Q, Ouyang S. Direct and indirect genotoxicity of graphene family nanomaterials on DNA-a review. Nanomaterials. 2021;11(11):2889. doi:10.3390/nano11112889
4. He K, Zeng Z, Chen A, et al. Advancement of Ag-graphene based nanocomposites: an overview of synthesis and its applications. Small. 2018;14(32):e1800871. doi:10.1002/smll.201800871
5. Liao C, Li Y, Tjong SC. Bactericidal and cytotoxic properties of silver nanoparticles. Int J Mol Sci. 2019;20(2):449. doi:10.3390/ijms20020449
6. Lu C, Lv Y, Kou G, et al. Silver nanoparticles induce developmental toxicity via oxidative stress and mitochondrial dysfunction in zebrafish (Danio rerio). Ecotoxicol Environ Saf. 2022;243:113993. doi:10.1016/j.ecoenv.2022.113993
7. Juling S, Böhmert L, Lichtenstein D, et al. Comparative proteomic analysis of hepatic effects induced by nanosilver, silver ions and nanoparticle coating in rats. Food Chem Toxicol. 2018;113:255–266. doi:10.1016/j.fct.2018.01.056
8. Rezvani E, Rafferty A, McGuinness C, Kennedy J. Adverse effects of nanosilver on human health and the environment. Acta Biomater. 2019;94:145–159. doi:10.1016/j.actbio.2019.05.042
9. Kim S, Ryu DY. Silver nanoparticle-induced oxidative stress, genotoxicity and apoptosis in cultured cells and animal tissues. J Appl Toxicol. 2013;33(2):78–89. doi:10.1002/jat.2792
10. Jarosz A, Skoda M, Dudek I, Szukiewicz D. Oxidative stress and mitochondrial activation as the main mechanisms underlying graphene toxicity against human cancer cells. Oxid Med Cell Longev. 2016;2016:5851035. doi:10.1155/2016/5851035
11. Hu J, Lin W, Lin B, et al. Persistent DNA methylation changes in zebrafish following graphene quantum dots exposure in surface chemistry-dependent manner. Ecotoxicol Environ Saf. 2019;169:370–375. doi:10.1016/j.ecoenv.2018.11.053
12. Di Ianni E, Møller P, Vogel UB, Jacobsen NR. Pro-inflammatory response and genotoxicity caused by clay and graphene nanomaterials in A549 and THP-1 cells. Mutat Res Genet Toxicol Environ Mutagen. 2021;872:503405. doi:10.1016/j.mrgentox.2021.503405
13. Tao R, Wang C, Zhang C, et al. Characterization, cytotoxicity and genotoxicity of graphene oxide and folate coupled chitosan nanocomposites loading polyprenol and fullerene based nanoemulsion against MHCC97H cells. J Biomed Nanotechnol. 2019;15(3):555–570. doi:10.1166/jbn.2019.2698
14. Pogribna M, Hammons G. Epigenetic effects of nanomaterials and nanoparticles. J Nanobiotechnology. 2021;19(1):2. doi:10.1186/s12951-020-00740-0
15. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi:10.1038/s41571-021-00585-y
16. Sierra MI, Valdés A, Fernández AF, Torrecillas R, Fraga MF. The effect of exposure to nanoparticles and nanomaterials on the mammalian epigenome. Int J Nanomedicine. 2016;11:6297–6306. doi:10.2147/IJN.S120104
17. Wong BSE, Hu Q, Baeg GH. Epigenetic modulations in nanoparticle-mediated toxicity. Food Chem Toxicol. 2017;109(Pt1):746–752. doi:10.1016/j.fct.2017.07.006
18. Yu J, Loh XJ, Luo Y, Ge S, Fan X, Ruan J. Insights into the epigenetic effects of nanomaterials on cells. Biomater Sci. 2020;8(3):763–775. doi:10.1039/c9bm01526d
19. Huang Y, Lü X, Lü X. Cytotoxic mechanism for silver nanoparticles based high-content cellomics and transcriptome sequencing. J Biomed Nanotechnol. 2019;15(7):1401–1414. doi:10.1166/jbn.2019.2785
20. Huang Y, Lü X, Lü X. Study of key biological pathways and important microRNAs involved in silver nanoparticles induced cytotoxicity based on microRNA sequencing technology. J Biomed Nanotechnol. 2018;14(12):2042–2055. doi:10.1166/jbn.2018.2643
21. Tao L, Chen X, Sun J, Wu C. Silver nanoparticles achieve cytotoxicity against breast cancer by regulating long-chain noncoding RNA XLOC_006390-mediated pathway. Toxicol Res. 2021;10(1):123–133. doi:10.1093/toxres/tfaa090
22. Yuan YG, Cai HQ, Wang JL, et al. Graphene oxide-silver nanoparticle nanocomposites induce oxidative stress and aberrant methylation in caprine fetal fibroblast cells. Cells. 2021;10(3):682. doi:10.3390/cells10030682
23. Yuan YG, Xing YT, Liu SZ, et al. Identification of circular RNAs expression pattern in caprine fetal fibroblast cells exposed to a chronic non-cytotoxic dose of graphene oxide-silver nanoparticle nanocomposites. Front Bioeng Biotechnol. 2023;11:1090814. doi:10.3389/fbioe.2023.1090814
24. Pan Y, Zhang W, Lin S. Transcriptomic and microRNAomic profiling reveals molecular mechanisms to cope with silver nanoparticle exposure in the ciliate Euplotes vannus. Environ Sci Nano. 2018;5(12):2921–2935. doi:10.1039/C8EN00924D
25. Ayala A, Muñoz MF, Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi:10.1155/2014/360438
26. Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830(5):3217–3266. doi:10.1016/j.bbagen.2012.09.018
27. Xu H, Liu Y, Cheng P, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39(1):283. doi:10.1186/s13046-020-01799-1
28. Barbasz A, Czyżowska A, Piergies N, Oćwieja M. Design cytotoxicity: the effect of silver nanoparticles stabilized by selected antioxidants on melanoma cells. J Appl Toxicol. 2022;42(4):570–587. doi:10.1002/jat.4240
29. McShan D, Ray PC, Yu H. Molecular toxicity mechanism of nanosilver. J Food Drug Anal. 2014;22(1):116–127. doi:10.1016/j.jfda.2014.01.010
30. Yousef HN, Ibraheim SS, Ramadan RA, Aboelwafa HR, Gonçalves-de-albuquerque CF. The ameliorative role of eugenol against silver nanoparticles-induced hepatotoxicity in male Wistar rats. Oxid Med Cell Longev. 2022;2022:3820848. doi:10.1155/2022/3820848
31. Yuan YG, Gurunathan S. Combination of graphene oxide-silver nanoparticle nanocomposites and cisplatin enhances apoptosis and autophagy in human cervical cancer cells. Int J Nanomedicine. 2017;12:6537–6558. doi:10.2147/IJN.S125281
32. Yuan YG, Wang YH, Xing HH, Gurunathan S. Quercetin-mediated synthesis of graphene oxide-silver nanoparticle nanocomposites: a suitable alternative nanotherapy for neuroblastoma. Int J Nanomedicine. 2017;12:5819–5839. doi:10.2147/IJN.S140605
33. Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol Histol. 2006;37(5–7):261–269. doi:10.1007/s10735-006-9047-4
34. Musolino E, Pagiatakis C, Serio S, et al. The Yin and Yang of epigenetics in the field of nanoparticles. Nanoscale Adv. 2022;4(4):979–994. doi:10.1039/d1na00682g
35. Huang Y, Lü X, Ma J. Toxicity of silver nanoparticles to human dermal fibroblasts on microRNA level. J Biomed Nanotechnol. 2014;10(11):3304–3317. doi:10.1166/jbn.2014.1974
36. Eom HJ, Chatterjee N, Lee J, Choi J. Integrated mRNA and micro RNA profiling reveals epigenetic mechanism of differential sensitivity of Jurkat T cells to AgNPs and Ag ions. Toxicol Lett. 2014;229(1):311–318. doi:10.1016/j.toxlet.2014.05.019
37. Oh JH, Son MY, Choi MS, et al. Integrative analysis of genes and miRNA alterations in human embryonic stem cells-derived neural cells after exposure to silver nanoparticles. Toxicol Appl Pharmacol. 2016;299:8–23. doi:10.1016/j.taap.2015.11.004
38. Ahamed M, Karns M, Goodson M, et al. DNA damage response to different surface chemistry of silver nanoparticles in mammalian cells. Toxicol Appl Pharmacol. 2008;233(3):404–410. doi:10.1016/j.taap.2008.09.015
39. Teodoro JS, Silva R, Varela AT, et al. Low-dose, subchronic exposure to silver nanoparticles causes mitochondrial alterations in Sprague-Dawley rats. Nanomedicine. 2016;11(11):1359–1375. doi:10.2217/nnm-2016-0049
40. Liu Y, Wen H, Wu X, et al. The bio-persistence of reversible inflammatory, histological changes and metabolic profile alterations in rat livers after silver/gold nanorod administration. Nanomaterials. 2021;11(10):2656. doi:10.3390/nano11102656
41. Shen L, Wang Q, Liu R, et al. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46(2):717–729. doi:10.1093/nar/gkx1224
42. Papaspyropoulos A, Lagopati N, Mourkioti I, et al. Regulatory and functional involvement of long non-coding RNAs in DNA double-strand break repair mechanisms. Cells. 2021;10(6):1506. doi:10.3390/cells10061506
43. Liu L, Jia Y, Zhang X, et al. Identification of the function and regulatory network of circ_009773 in DNA damage induced by nanoparticles of neodymium oxide. Toxicol In Vitro. 2022;78:105271. doi:10.1016/j.tiv.2021.105271
44. Spitzer N, Patterson KK, Kipps DW. Akt and MAPK/ERK signaling regulate neurite extension in adult neural progenitor cells but do not directly mediate disruption of cytoskeletal structure and neurite dynamics by low-level silver nanoparticles. Toxicol In Vitro. 2021;74:105151. doi:10.1016/j.tiv.2021.105151
45. Parnsamut C, Brimson S. Effects of silver nanoparticles and gold nanoparticles on IL-2, IL-6, and TNF-α production via MAPK pathway in leukemic cell lines. Genet Mol Res. 2015;14(2):3650–3668. doi:10.4238/2015.April.17.15
46. Zhao X, Rao Y, Liang J, et al. Silver nanoparticle-induced phosphorylation of histone H3 at serine 10 involves MAPK pathways. Biomolecules. 2019;9(2):78. doi:10.3390/biom9020078
47. Xue M, Momiji H, Rabbani N, et al. Frequency modulated translocational oscillations of Nrf2 mediate the antioxidant response element cytoprotective transcriptional response. Antioxid Redox Signal. 2015;23:613–629. doi:10.1089/ars.2014.5962
48. Yang Y, Ren J, Huang Q, et al. CircRNA expression profiles and the potential role of CircZFP644 in mice with severe acute pancreatitis via sponging miR-21-3p. Front Genet. 2020;11:206. doi:10.3389/fgene.2020.00206
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.