Back to Journals » Infection and Drug Resistance » Volume 14
Multiple Cross Displacement Amplification Linked with Nanoparticles-Based Lateral Flow Biosensor in Screening of Hepatitis B Virus in Clinical Application
Authors Chen X, Zhou Q , Dong S , Wang S, Liu R, Wu X, Li S
Received 17 December 2020
Accepted for publication 9 March 2021
Published 25 March 2021 Volume 2021:14 Pages 1219—1229
DOI https://doi.org/10.2147/IDR.S297645
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Xu Chen,1,2,* Qingxue Zhou,3,* Shilei Dong,4 Shuoshi Wang,2 Rui Liu,2 Xueli Wu,2 Shijun Li5
1The Second Clinical College, Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, 550003, People’s Republic of China; 2Central Laboratory of the Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine, Guiyang, Guizhou, 550003, People’s Republic of China; 3Clinical Laboratory, Hangzhou Women’s Hospital, Hangzhou, Zhejiang, 310008, People’s Republic of China; 4Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou, Zhejiang, 310013, People’s Republic of China; 5Laboratory of Bacterial Infectious Disease of Experimental Centre, Guizhou Provincial Centre for Disease Control and Prevention, Guiyang, Guizhou, 550004, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shijun Li
Guizhou Provincial Centre for Disease Control and Prevention, 115 Bageyan Road, Guiyang, Guizhou, 550004, People’s Republic of China
Tel/Fax +86 851 86824290
Email [email protected]
Xu Chen
The Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine, 83 Feishan Road, Guiyang, Guizhou, 550003, People’s Republic of China
Tel/Fax +86 851 85514432
Email [email protected]
Background: Hepatitis B virus (HBV) is a common pathogen that predominantly causes severe liver disease, and remains one of a huge challenge worldwide, especially in many resource-constrained areas. Developing a low-cost, sensitive, specific, and rapid approach for screening HBV is critical for its treatment and prevention. In the current study, a novel molecular detection approach, multiple cross displacement amplification (MCDA) coupled with polymer nanoparticle-based lateral flow biosensor (MCDA-LFB), was applied for detection of HBV in blood samples.
Methods: HBV standard substance and clinical donor serum samples were collected and used for the establishment and confirmation of the HBV-MCDA-LFB assay. A set of 10 MCDA primers was designed according to HBV-specific gene S. The HBV-MCDA-LFB assay conditions, including genomic template concentration, MCDA reaction temperature and time were optimized. The sensitivity and specificity of the HBV-MCDA -LFB assay were evaluated in this report. The HBV-MCDA-LFB assay was applied to detect the HBV agent from clinical samples.
Results: The HBV-MCDA primers based on the S gene were valid for establishment of MCDA assay. The HBV-MCDA reaction with optimized conditions could be carried out at a constant temperature 64°C for 35 min. The whole process, including sample preparation (5 min), genomic template extraction (∼ 30 min), MCDA amplification (35 min), and LFB reading (∼ 2 min), could be completed within 80 min. The sensitivity of this assay was 5 IU per reaction. The specificity was 100% for HBV-MCDA-LFB assay.
Conclusion: These results confirmed that the HBV-MCDA-LFB is a low-cost, sensitive, specific, simple, and rapid method for detecting HBV agents. This technique has great potential to develop a point-of-care testing (POCT) method in clinical practice, especially in endemic and resource-constrained regions.
Keywords: hepatitis B virus, HBV, multiple cross displacement amplification, MCDA, lateral flow biosensor, LFB, rapid detection, molecular detection
Introduction
Hepatitis B virus (HBV), an enveloped and partially double-stranded DNA genome virus, is a common pathogen that predominantly causes severe liver diseases such as liver cirrhosis, liver failure, and hepatocellular carcinoma (HCC).1–3 Currently, HBV infection remains one a major health challenge worldwide. According to a 2017 World Health Organization (WHO) report, an estimated 257 million people were living with chronic HBV infection, and 15–40% of them will develop into liver cirrhosis or HCC.4 Currently there is no effective antiviral therapy which can eradicate HBV infection,5 vaccination has become the only valid strategy for preventing HBV infection. Unfortunately, HBV vaccine is not suitable for some individuals, such as those with type I diabetes mellitus, human immunodeficiency virus (HIV) infection, and chronic kidney disease.5,6 Thus, it is important to develop a sensitive, specific, and rapid assay for monitoring HBV at the early stage of infection, which could help in addressing the burden of HBV infection in local health care systems, and provide a guideline to the government and clinicians in designing preventive and therapeutic strategies.
Currently, enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) related technologies are the most commonly used methods for HBV identification in clinical diagnostic application.7,8 Immunoassay is based on a serological response targeting the HBV antigens or antibodies, which can present better accuracy.9 However, the sensitivity of the ELISA method is low at the early stages of HBV infection.9,10 In recent decades, many molecular methods, including PCR, multiplex PCR, and quantitative PCR (qPCR), have been widely applied for detecting HBV in clinical practice.11,12 Nevertheless, such techniques depend on expensive thermo-cycling instruments to separate DNA strands and amplify the target genes that may not be available in many resource-constrained regions.11 Herein, developing a cost-effective, sensitive, specific, simple, and rapid technique for identification of HBV is necessary for the prevention and follow-up treatment of HBV infected patients.
To overcome the drawbacks of such traditional detections, the multiple cross displacement amplification (MCDA) technique is a novel, low-cost, sensitive, and rapid isothermal amplification technology similar to LAMP, but with more specificity and sensitivity in microorganism assay,13 which has been applied to detect several pathogens such as Mycobacterium tuberculosis, Neisseria meningitides, and Listeria monocytogenes.14–16 Previous studies suggested that MCDA amplification products could been analyzed by various technologies, including agarose gel, turbidimetry, and colorimetric indicators (malachite green reagent).17 However, both agarose gel and turbidimetry need specific facilities.14 Malachite green reagent is simple to operation, but it is easy to ambiguous when the concentration of the amplicons was low.18 Besides, all of these detection methods are not specific for target gene and are easy to cause false-positive result. To overcome these defects, polymer nanoparticle-based lateral flow biosensor (LFB) was successfully designed and applied to analyze MCDA products, In comparison with such methods, LFB is relatively specific, objective, cost-effective, and simple.
In the present study, MCDA amplification combined with LFB detector (MCDA-LFB) was developed for cost-effective, sensitive, specific, rapid, simple, and visual identification of HBV carrying the S gene,7 which appeared to be uniquely present in HBV as it showed no homology with other microbial genomes at GenBank by BLAST searches. The detection performance was analyzed with HBV nucleic acid (HBV DNA) standard substance and clinical serum samples.
Materials and Methods
Ethics Statement
The study was approved by the Human Ethics Committee of Hangzhou Women’s Hospital (Approval No. [2020]-A-10-9) complied with the Declaration of Helsinki. Before our team obtained the samples/isolates and conducted study, any personal identifiers of the suspected HBV-infected patients had been removed by the monitoring stations. So the patient informed consents were waived by the Human Ethics Committee of Hangzhou Women’s Hospital.
Standard Panels and Clinical Samples Preparation
In the current study, HBV (Code No. GBW09150), HIV (Code No. GBW (E) 090272), and Hepatitis C Virus (HCV) (Code No. GBW (E) 090118) nucleic acid standard substances were obtained from the Chinese Academy of Metrology. The 136 clinical serum samples were collected from suspected HBV infected patients at the Hangzhou Women’s Hospital (Hangzhou, China) during January 2020 to September 2020, other non-HBV pathogens were used in this study were shown in Table 1.
 |
Table 1 Pathogens Used in the Current Study |
Materials and Reagents
Genomic DNA/RNA nucleic acid extraction kits were obtained from Tianlong Technology Co., Ltd (Xian, China). Colorimetric indicator (Malachite Green, MG) and universal isothermal amplification kits were purchased from Bei-Jing HaiTaiZhengYuan. Co., Ltd. (Beijing, China). Biotin-BSA (biotinylated bovine serum albumin) and Anti-FAM (rabbit anti-fluorescein antibody) were obtained from Abcam. Co., Ltd. (Shanghai, China). Dye streptavidin-coated polymer nanoparticles (Crimson red) were obtained from Bangs Laboratories, Inc. (Indiana, USA). Polymer nanoparticle-based lateral flow biosensor (LFB) materials, including sample pad, conjugate pad, absorbent pad, nitrocellulose membrane (NC) and backing card were obtained from Jie-Yi Biotechnology. Co., Ltd. (Shanghai, China). HBV qPCR diagnosis kits were purchased from Da An Gene Co., Ltd. (Guangzhou, China). The purity and concentration of nucleic acid was analyzed with Nano-Drop ND-2000 (Beijing, China) at A260/280.
Design of HBV-MCDA Primers
According to the mechanism of MCDA, five pairs of special primers based on S gene (Genbank Accession No. AB809557.1) of HBV were designed using primer Explorer V5 and PRIMER PREMIER 5.0 software (Eiken Chemical, Japan). Hairpin structures and hybrids of these primers were analyzed with Integrated DNA Technologies design tool. The specificity of HBV-MCDA primers were verified using BLAST analysis tool. The locations of MCDA primers are shown in Figure 1, and the sequences of these primers are shown in Table 2. All of the oligomers were synthesized and purified by TsingKe Biotech Co., Ltd. (Beijing, China) with high performance liquid chromatography purification grade.
 |
Table 2 The HBV-MCDA Primers Used in the Current Study |
Preparation of Nanoparticle-Based Lateral Flow Biosensor
Polymer nanoparticle-based LFB used in this study was designed as previous report.19 In brief, the biosensor was composed of four sections, including sample pad, conjugate pad, NC membrane, and absorbent pad. Dye streptavidin-coated polymer nanoparticles were laminated in the conjugate pad. Anti-FAM and biotin-BSA were conjugated onto the NC membrane for test line (TL) and control line (CL), respectively. The prepared LFBs were preserved with a desiccant gel at room temperature.
Standard MCDA Reactions
The HBV-MCDA amplification was performed in a one-step reaction of 25 μL mixture containing 0.4 μM of F1 and F2; 1.2 μM of D1*, D2, R1 and R2; 0.8 μM of C1* and C2; 1.2 μM of CP1 and CP2 (cross primers); 12.5 μL of 2×reaction mix; 1 μL of Bst DNA polymerase (10 U); 1 μL (8 U) of AMV Reverse Transcriptase (only used for RNA temperate); and 1 μL nucleic acid template. The mixtures were heated at 63°C for 1 h. HBV nucleic acid standard substances were obtained from Chinese Academy of Metrology, non-HBV nucleic acid, including HCV and HIV were used as negatives controls (NCs). The HIV and HCV nucleic acid standard substances were obtained from the Chinese Academy of Metrology, and the concentration was 1×105 IU per reaction. Double distilled water (DW) was used as the template in blank control (BC). Each reaction was tested at least three times.
HBV-MCDA Products Detection
Three MCDA detection methods, including Colorimetric indicator (Malachite Green [MG]), real-time turbidity (LA-500), and LFB were employed for determining and verifying the HBV-MCDA products, respectively. For colorimetric indicator detection, the color changed from colorless to light green in reaction system, indicating positive amplification products, but the color remains colorless in negative control (NC). For real-time turbidity method, turbidity>0.1 was considered as positive results. For LFB detection, both the CL and TL appeared simultaneously, indicating positive results, but in negative amplification only the CL could be observed.
Optimization of the HBV-MCDA Reaction Temperature
In the MCDA reaction system, temperature was essential for influencing amplification efficiency. The reaction temperature was optimized ranging from 60–67°C (with 1°Cintervals). The HBV-MCDA amplicons were determined with real-time turbidity (LA-500). Turbidity > 0.1 was considered as positive result.
Analytical Sensitivity of HBV-MCDA-LFB Assay
To test the sensitivity of HBV-MCDA-LFB assay, HBV DNA standard substance was 10-fold serial diluted from 5.0×103 to 5.0×10−2 IU. The HBV-MCDA amplification was carried out at the optimized temperature, and the products were monitored with MG and LFB. The sensitive of HBV-MCDA-LFB assay was confirmed as the last dilution of each positive test. Three replicates of each dilution were tested.
Optimization of the Amplification Time for HBV-MCDA-LFB Assay
In the current study, the reaction time of the HBV-MCDA was also optimized. Four reaction times (25, 35, 45, 55 min) were tested and compared. The HBV-MCDA reaction was carried out as described above, and the amplification products were monitored using LFB. Each amplification time was tested at least three times.
Specificity Analysis of HBV-MCDA-LFB Assay
The specificity of the HBV-MCDA-LFB assay was examined using nucleic acid (at least 10 ng per reaction) from various pathogens, including HBV nucleic acid standard substance, 18 HBV-positive clinical serum samples and 20 non-HBV pathogens (Table 1). DW was used as a BC. All of the MCDA results were monitored with an LFB and confirmed at least three times.
Examination of the Feasibility of HBV-MCDA-LFB Assay Using Clinical Samples
For verifying the feasibility of HBV-MCDA-LFB assay, the optimized HBV-MCDA-LFB detection system was directly evaluated using clinical samples in parallel with qPCR diagnosis method. One hundred and thirty-six suspected HBV-infection serum samples were collected from Hangzhou Women’s Hospital (Hangzhou, China). The HBV-LAMP-LFB operation as described above, and qPCR diagnosis was carried out using commercial real-time TaqMan PCR Kit (Da An Gene Co., Ltd. China) according to the manufacturer’s instructions. The results of HBV-MCDA-LFB were compared with that of qPCR assay.
Results
Demonstration of HBV-MCDA Products
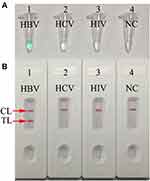
To test the valid of the MCDA primers (Table 2), for HBV assay. HBV-MCDA mixtures were carried out at a constant temperature of 63°C for 1 h using HBV nucleic acid standard substance as template. Then, the amplification products were monitored by two different methods, including MG and LFB. The results showed that the positive results were observed when the nucleic acid from HBV, but not with HCV, HIV, and the BC (Figure 2A and B). Hence, the HBV-MCDA primers for S gene detection in the current study were valid for the establishment of HBV-MCDA-LFB assay.
Optimal HBV-MCDA Assay Temperature
To confirm the optimal temperature for isothermal amplification, the HBV-MCDA reaction was performed from 60 to 67 °C with 100 IU/reaction of HBV nucleic acid as template. All of the reactions were monitored with real-time turbidity (LA-500). The kinetic data in Figure 3 showed that the faster amplifications of HBV-MCDA were observed at the temperature range from 64 to 67°C. Therefore, the temperature of 64°C was considered as optimal amplification temperature for the subsequent HBV-MCDA assay in the current study.
Sensitivity of the HBV-MCDA-LFB Assay
The limits of detection (LoD) of HBV-MCDA-LFB was determined with serially dilutions of HBV nucleic acid standard substance (5.0×103 IU, 5.0×102 IU, 5.0×101 IU, 5 IU, 0.5 IU, 0.05 IU). The MCDA reaction was carried out as described above, and the products were monitored using MG reagent and LFB technique. The results demonstrated that the LoD of HBV-MCDA-LFB was 5 IU (Figure 4A) using MG method, which was complete consistent with LFB analysis (Figure 4B).
Optimal Amplification Time for HBV-MCDA-LFB Assay
In order to optimize amplification time for the HBV-MCDA-LFB assay, four amplification times (25, 35, 45, and 55 min) were tested at 64°C, respectively. The amplification products were analyzed with LFB. The results confirmed that the LoD level of HBV genomic DNA (5 IU per reaction) was tested when the amplification last 35, 45, and 55 min (Figure 5). Therefore, 35 min was considered an appropriate amplification time for HBV-MCDA-LFB assay.
Specificity Analysis of the HBV-MCDA-LFB Assay
The specificity of HBV-MCDA-LFB assay was identified with HBV nucleic acid standard substance, 18 HBV-positive clinical samples and 20 non-HBV pathogens (Table 1). The MCDA-LFB detection was performed as the optimal conditions confirmed above. The results showed that only the genomic DNA from HBV agents presented positive results, while other non-HBV pathogens and BC showed negative results (Figure 6). Therefore, the specification of MCDA-LFB assay established in the current study was 100% for HBV assay.
Evaluation of Feasibility of HBV-MCDA-LFB Assay in Clinical Specimens
To further verify whether the HBV-MCDA-LFB technique could be applied to clinical diagnosis, 136 suspected HBV-infection serum samples were simultaneously detected with HBV-MCDA-LFB and qPCR. Of the 136 suspected HBV-infection samples, 89 samples were identified positive by qPCR (IU > 30). These HBV positive samples have also been identified with HBV-MCDA-LFB. More importantly, 4 of the 47 “negative samples” (10~20 IU) were tested by HBV-MCDA-LFB technique, and presented positive results (Table 3). These results indicated that the HBV-MCDA-LFB technology was more sensitive than the qPCR, and could be used as an advanced tool to detect HBV agent, especially suit for screening of the hepatitis B patients at the initial stage of infection.
 |
Table 3 Comparison of qPCR and HBV-MCDA-LFB Methods to Detect HBV Agent in Clinical Samples |
Discussion
HBV is one of the most dangerous and prevalent agents that causes acute and chronic hepatic infections and liver cancer in humans.20,21 Generally, transmission of HBV occurs mainly due to blood transfusion or vertical transmission.22 More importantly, HBV is often co-infected with HCV and HIV, resulting in a significant public health and socioeconomic burden all over the world, especially in underprivileged countries and regions.23,24 Thus, early and rapid detection of HBV in suspected patients is essential to control infection by this pathogen. To date, ELISA and real-time PCR have been developed to identify HBV in clinical samples.9 However, the sensitivity of the ELISA is too low for screening HBV infection at early stage.25 In recent years, qPCR has become the standard assay for HBV screening in clinical setting for its wide dynamic range of viral detection (range from 10 to 108 IU per reaction), specificity, and pollution control.6,26,27 Comparing of enzyme immunoassay assay, qPCR could reduce time for identification of HBV after exposure from an average of approximately 32 days (HBsAg marker) to 15 days (HBV DNA marker).28,29 Nevertheless, this assay is time-consuming, and need expensive facilities and trained experts. It is not readily affordable particularly in resource-limitation regions. In the current study, we first report a simple, low-cost, sensitive and specific MCDA-LFB assay to identify HBV agent.
MCDA is a novel isothermal amplification method, which was first reported by Wang et al in 2015.30 It involves only one enzyme, a polymerase with strand displacement capability and is isothermal, operating at a single temperature (between 60–68°C) throughout the reaction. The isothermal amplification of specific nucleic-acid sequences is achieved by employing a set of 10 primers spanning 10 distinct regions of target fragment, which are designated as displacement primers (F1 and F2), cross primers (CP1 and CP2) and amplification primers (D1, D2, C1, C2, R1 and R2). Recent development of nanobiotechnology has given birth to advanced techniques for diagnosis application.31 Polymer nanoparticles have high surface-to-volume ratio, biocompatible, high adsorption and reactive capacity, easy to synthesis and manipulate. Because of these properties, polymer nanoparticles are the most appropriate nanomaterial used as biosensor.32 In the current study, the MCDA linked with polymer nanoparticle-based LFB approach targeting S gene was established for identifying HBV agent. Five pairs of HBV-MCDA primers, which specially recognized 10 regions of the S gene, were devised according to MCDA mechanism.30 The specificity of HBV-MCDA-LFB assay was powerfully verified with nucleic acid extracted from HBV clinical samples and other pathogens. All of the HBV samples displayed positive outcomes, but non-HBV agents showed negative results (Figure 6). Thus, the HBV-MCDA-LFB assay displayed a high level of specificity for the detection of HBV agent. In addition to its excellent specificity, the newly developed HBV-MCDA-LFB method was able to detect 5 IU genomic DNA per reaction. Importantly, we also successfully applied the HBV-MCDA-LFB assay to clinical serum samples. The suspected HBV-infection samples were simultaneously detected using qPCR and HBV-MCDA-LFB. The qPCR diagnosis was carried out using commercial real-time TaqMan PCR Kit (Da An Gene Co., Ltd. China). According to the manufacturer’s instructions, the concentrations of HBV less than 30 IU will be considered as negative results. In the current study, 4 “negative” clinical samples (HBV concentrations range from 10 to 20) were tested with HBV-MCDA-LFB, and presented positive results (Table 3). It is indicated that HBV-MCDA-LFB is more sensitive than qPCR.
In this report, polymer nanoparticles as carrier materials for preparation of nanoparticle-based LFB. The LFB designed in this study could visually detect the MCDA products for labeling with anti-FAM and BSA-biotin on the LFB strips. The two crimson red bands CL and TL appeared in the LFB strip indicating positive results, while the negative results only appeared CL in the biosensor. Although the real-time turbidity and colorimetric indicator (MG reagent) could identify HBV-MCDA products in the present study, real-time turbidimeter is laborious and requires specific, dedicated equipment. Colorimetric indicator is simple, but it could somewhat ambiguous when the concentration of the amplicon was low. Hence, LFB was considered as a promising candidate to detect the HBV-MCDA products.
Compared with qPCR assay, the HBV-MCDA-LFB technique is more time-saving and cost-saving. Firstly, the entire detection process, including sample preparation (taking venous blood and separating serum through centrifugation) (~5 min), genomic DNA template preparation (~30 min), MCDA amplification (35 min) and LFB analysis (~2 min), could be completed within 80 min. However, the qPCR requires 2~3 h during the whole process. On the other hand, the HBV-MCDA-LFB is less dependent on expensive apparatuses and reagents. As an isothermal amplification assay, a simple heating instrument (such as water-bath or heater) is sufficient for MCDA. The total cost of each test, including genomic DNA template preparation (~ $1 USD), MCDA amplification (~ $3.5 USD) and LFB identifying (~ $2 USD), is estimated to be $ 6.5 USD, which is cheaper than real-time PCR technique. More importantly, the HBV-MCDA-LFB technology has great potential to develop a POCT method for screening HBV-suspected patients in clinical setting.
The HBV-MCDA-LFB method also has some limitations in the current study. Firstly, the newly MCDA-LFB detection is a qualitative determination for HBV, and could not quantify the amount of pathogen in samples. A more precise study will be designed for quantification of the amount of HBV in clinical samples with MCDA-LFB assay. Secondly, HBV-MCDA amplifications must be taken out from the reaction tube for LFB detection, carry-over contamination is a great concern for this method. But the possibility of laboratory contamination is very low. First, the LFB is very sensitivity that just needs 0.2~0.5μL MCDA products for detection, and then the LFB platform has high adsorptive capacity that can rapidly remove MCDA products from sample region to reaction regions. So, it is hard to form aerosols. Second, after completion of detection, spraying 10~15% sodium hypochlorite solution and 70% ethanol is also an effective way to overcome nucleic acid contamination of laboratory. In the present study, the false positive results have not appeared in our laboratory.
In conclusion, we established a sensitive, specific, simple, rapid, and low cost HBV-MCDA-LFB assay based on the S gene for screening HBV agent in clinical practice. The LoD of the advanced assay for HBV detection was as low as 5 IU. Compared with other molecular methods, such as PCR and qPCR, the HBV-MCDA-LFB technique is more time-saving and cost-saving, and does not require expensive apparatus, which has the great potential to be an important POCT method for HBV surveillance and detection at the early stage of infection, especially in resource-limited regions of the world.
Abbreviations
WHO, World Health Organization; NCBI, National Centre for Biotechnology Information; HBV, hepatitis B Virus; HCV, Hepatitis C Virus; HIV, human immunodeficiency virus; GZCDC, Guizhou Provincial Center for Disease Control and Prevention; 2nd GZUTCM, The Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine; FAM, 6-carboxy-fluorescein; MG, malachite green; nt, nucleotide; mer, monomeric unit; DW, distilled water; POCT, Point-of-Care testing; CL, control line; TL, test line; NC, negative control; BC, blank control.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We thank all the medical workers in the Hangzhou Women’s Hospital, The Second Affiliated Hospital, Guizhou University of Traditional Chinese Medicine, and Guizhou Provincial Center for Disease Control and Prevention for their cooperation in the current study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Program of Scientific and Technological Project in Guizhou Province (Grant No. Qian Ke He [2020]4Y184 and [2019]1186), the Scientific and Technological in Guiyang City (Grant No. Zhu Ke He [2020]-10-5), the Scientific and Technological of 2nd GZUTCM (Grant No. GZEYK [2020] 29), the Program of the High-Level Creative Talents Cultivation in Guizhou Province (Qian Ke He (2016)4021), the Program of Scientific and Technological Innovation Team of Guizhou Province (Grant No. Qian Ke He Platform talent [2018]5606), the Public Welfare Technology Research Program in Zhejiang Province (Grant No. LGF21H190001).
Disclosure
All of the authors declare that there are no competing interests associated with the manuscript.
References
1. Joshi SS, Coffin CS. Hepatitis B and pregnancy: virologic and immunologic characteristics. Hepatol Commun. 2020;4(2):157–171. doi:10.1002/hep4.1460
2. Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49(5 Suppl):S13–21. doi:10.1002/hep.22881
3. Wu CC, Chen YS, Cao L, et al. Hepatitis B virus infection: defective surface antigen expression and pathogenesis. World J Gastroenterol. 2018;24(31):3488–3499. doi:10.3748/wjg.v24.i31.3488
4. World Health Organization. Global hepatitis report. Geneva: WHO; 2017
5. Wang FJ, Lu LL, Yu CS, et al. Development of a novel DNA sequencing method not only for hepatitis B virus genotyping but also for drug resistant mutation detection. BMC Med Genomics. 2013;6 Suppl 1(Suppl 1):S15. doi:10.1186/1755-8794-6-S1-S15
6. Zhao JR, Bai YJ, Zhang QH, et al. Detection of hepatitis B virus DNA by real-time PCR using TaqMan-MGB probe technology. World J Gastroenterol. 2005;11(4):508–510. doi:10.3748/wjg.v11.i4.508
7. Jia SR, Wang F, Li F, et al. Rapid detection of hepatitis B virus variants associated with lamivudine and adefovir resistance by multiplex ligation-dependent probe amplification combined with real-time PCR. J Clin Microbiol. 2014;52(2):460–466. doi:10.1128/JCM.02554-13
8. Liu YP, Yao CY. Rapid and quantitative detection of hepatitis B virus. World J Gastroenterol. 2015;21(42):11954–11963. doi:10.3748/wjg.v21.i42.11954
9. Ocana S, Casas ML, Buhigas I, et al. Diagnostic strategy for occult hepatitis B virus infection. World J Gastroenterol. 2011;17(12):1553–1557. doi:10.3748/wjg.v17.i12.1553
10. Ghosh M, Nandi S, Dutta S, et al. Detection of hepatitis B virus infection: a systematic review. World J Hepatol. 2015;7(23):2482–2491. doi:10.4254/wjh.v7.i23.2482
11. Villar LM, Cruz HM, Barbosa JR, et al. Update on hepatitis B and C virus diagnosis. World J Virol. 2015;4(4):323–342. doi:10.5501/wjv.v4.i4.323
12. Francisca SJ, Daniel MM, Nora HRM, et al. Effectiveness of PCR primers for the detection of occult hepatitis B virus infection in Mexican patients. PLoS One. 2018;13(10):e0205356. doi:10.1371/journal.pone.0205356
13. Li SJ, Liu Y, Wang Y, et al. Rapid detection of Brucella spp. and elimination of carryover using multiple cross displacement amplification coupled with nanoparticles-based lateral flow biosensor. Front Cell Infect Microbiol. 2019;9:78. doi:10.3389/fcimb.2019.00078
14. Jiao WW, Wang Y, Wang GR, et al. Development and clinical validation of multiple cross displacement amplification combined with nanoparticles-based biosensor for detection of Mycobacterium tuberculosis: preliminary results. Front Microbiol. 2019;10:2135. doi:10.3389/fmicb.2019.02135
15. Li SJ, Liu CT, Liu Y, et al. Development of a multiple cross displacement amplification combined with nanoparticles-based biosensor assay to detect Neisseria meningitides.. Infect Drug Resist. 2019;12:2077–2087. doi:10.2147/IDR.S210735
16. Wang Y, Li H, Wang Y, et al. Development of multiple cross displacement amplification label-based gold nanoparticles lateral flow biosensor for detection of Listeria monocytogenes.. Int J Nanomedicine. 2017;12:473–486. doi:10.2147/IJN.S123625
17. Zhao F, Niu L, Yan LL, et al. Establishment and application of Multiple Cross Displacement Amplification Coupled With Lateral Flow Biosensor (MCDA-LFB) for Visual and rapid detection of Candida albicans in clinical samples. Front Cell Infect Microbiol. 2019;9:102. doi:10.3389/fcimb.2019.00102
18. Niu L, Zhao F, Chen JL, et al. Isothermal amplification and rapid detection of Klebsiella pneumoniae based on the multiple cross displacement amplification (MCDA) and gold nanoparticle lateral flow biosensor (LFB). PLoS One. 2018;13(10):e0204332. doi:10.1371/journal.pone.0204332
19. Chen X, Ma K, Yi X, et al. A novel detection of Enterococcus faecalis using multiple cross displacement amplification linked with gold nanoparticle lateral flow biosensor. Infect Drug Resist. 2019;12:3771–3781. doi:10.2147/IDR.S235325
20. Jain S, Su YH, Su YP, et al. Characterization of the hepatitis B virus DNA detected in urine of chronic hepatitis B patients. BMC Gastroenterol. 2018;18(1):40. doi:10.1186/s12876-018-0767-1
21. Wu IC, Liu WC, Chang TT. Applications of next-generation sequencing analysis for the detection of hepatocellular carcinoma-associated hepatitis B virus mutations. J Biomed Sci. 2018;25(1):51. doi:10.1186/s12929-018-0442-4
22. Said ZN. An overview of occult hepatitis B virus infection. World J Gastroenterol. 2011;17(15):1927–1938. doi:10.3748/wjg.v17.i15.1927
23. Blackard JT, Sherman KE. Hepatitis B virus (HBV) reactivation-The potential role of direct-acting agents for hepatitis C virus (HCV). Rev Med Virol. 2018;28(4):e1984. doi:10.1002/rmv.1984
24. Leoni MC, Ustianowski A, Farooq H, et al. HIV, HCV and HBV: a review of parallels and differences. Infect Dis Ther. 2018;7(4):407–419. doi:10.1007/s40121-018-0210-5
25. Portilho MM, Nabuco LC, Villela-Nogueira CA, et al. Detection of occult hepatitis B in serum and oral fluid samples. Mem Inst Oswaldo Cruz. 2018;113(1):62–65. doi:10.1590/0074-02760170071
26. Shen XX, Qiu FZ, Shen LP, et al. A rapid and sensitive recombinase aided amplification assay to detect hepatitis B virus without DNA extraction. BMC Infect Dis. 2019;19(1):229. doi:10.1186/s12879-019-3814-9
27. Coffin CS, Zhou K, Terrault NA. New and old biomarkers for diagnosis and management of chronic hepatitis B virus infection. Gastroenterology. 2019;156(2):355–368.e3. doi:10.1053/j.gastro.2018.11.037
28. Song JE, Kim DY. Diagnosis of hepatitis B. Ann Transl Med. 2016;4(18):338. doi:10.21037/atm.2016.09.11
29. Yao CY, Fu WL. Biosensors for hepatitis B virus detection. World J Gastroenterol. 2014;20(35):12485–12492. doi:10.3748/wjg.v20.i35.12485
30. Wang Y, Wang Y, Ma AJ, et al. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci Rep. 2015;5(1):11902. doi:10.1038/srep11902
31. Banerjee R, Jaiswal A. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst. 2018;143(9):1970–1996. doi:10.1039/C8AN00307F
32. Quesada-González D, Merkoçi A. Nanoparticle-based lateral flow biosensors. Biosens Bioelectron. 2015;73:47–63. doi:10.1016/j.bios.2015.05.050
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.