Back to Journals » International Journal of Nanomedicine » Volume 11
Multifunctional selenium nanoparticles as carriers of HSP70 siRNA to induce apoptosis of HepG2 cells
Authors Li Y , Lin Z, Zhao M, Xu T, Wang C, Xia H, Wang H, Zhu B
Received 4 April 2016
Accepted for publication 28 May 2016
Published 5 July 2016 Volume 2016:11 Pages 3065—3076
DOI https://doi.org/10.2147/IJN.S109822
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Lei Yang
Yinghua Li,1 Zhengfang Lin,1 Mingqi Zhao,1 Tiantian Xu,1 Changbing Wang,1 Huimin Xia,1,* Hanzhong Wang,2,* Bing Zhu1,*
1Guangzhou Women and Children’s Medical Center, Guangzhou, Guangdong, 2State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, People’s Republic of China
*These authors contributed equally to this work
Abstract: Small interfering RNA (siRNA) as a new therapeutic modality holds promise for cancer treatment, but it is unable to cross cell membrane. To overcome this limitation, nanotechnology has been proposed for mediation of siRNA transfection. Selenium (Se) is a vital dietary trace element for mammalian life and plays an essential role in the growth and functioning of humans. As a novel Se species, Se nanoparticles have attracted more and more attention for their higher anticancer efficacy. In the present study, siRNAs with polyethylenimine (PEI)-modified Se nanoparticles (Se@PEI@siRNA) have been demonstrated to enhance the apoptosis of HepG2 cells. Heat shock protein (HSP)-70 is overexpressed in many types of human cancer and plays a significant role in several biological processes including the regulation of apoptosis. The objective of this study was to silence inducible HSP70 and promote the apoptosis of Se-induced HepG2 cells. Se@PEI@siRNA were successfully prepared and characterized by various microscopic methods. Se@PEI@siRNA showed satisfactory size distribution, high stability, and selectivity between cancer and normal cells. The cytotoxicity of Se@PEI@siRNA was lower for normal cells than tumor cells, indicating that these compounds may have fewer side effects. The gene-silencing efficiency of Se@PEI@siRNA was significantly much higher than Lipofectamine 2000@siRNA and resulted in a significantly reduced HSP70 mRNA and protein expression in cancer cells. When the expression of HSP70 was diminished, the function of cell protection was also removed and cancer cells became more sensitive to Se@PEI@siRNA. Moreover, Se@PEI@siRNA exhibited enhanced cytotoxic effects on cancer cells and triggered intracellular reactive oxygen species, and the signaling pathways of p53 and AKT were activated to advance cell apoptosis. Taken together, this study provides a strategy for the design of an anticancer nanosystem as a carrier of HSP70 siRNA to achieve synergistic cancer therapy.
Keywords: selenium nanoparticles, siRNA, polyethylenimine, reactive oxygen species, apoptosis, anticancer
Introduction
Small interfering RNA (siRNA) is a synthetic double-stranded RNA composed of 21–23 nucleotides, which leads to a reduction in the level of messenger RNA (mRNA) by suppressing the expression of the target gene.1,2 siRNA holds great promise as a powerful research tool and represents a novel method for cancer therapy.3,4 However, the therapeutic applications of siRNA have been limited by its strong negative charge, instability under nuclease, and low transfection efficiency, which lead to rapid enzymatic degradation and inability to cross the cell membrane.5,6 Recently, different kinds of gene vectors, including lipids, peptides, iron oxide, silica, gold nanoparticles, and cationic polymers have been investigated widely as carriers for siRNA delivery.7–9 The major challenge of effective delivery systems is to avoid potentially toxic side effects and low transfection efficiency. As one of the most traditional polycationic polymers, polyethylenimine (PEI) has been developed to protect the siRNA release from endosomes to achieve gene delivery.10–13 Heat shock proteins (HSPs) are named according to their molecular weight, as three major members of the HSP family, HSP27, HSP70, and HSP90 are related to human cancer.14 HSP70 is normally at low levels, but it is inducible under protein damage and overexpressed in different tumor types.15 Because of its ability to induce antitumor immune responses, HSP70 provides an innovative approach for cancer immunotherapy.16 Serum level of HSP70 could be a sensitive marker for the diagnosis of hepatocellular carcinoma.17 As a potential target for tumor treatment, many HSP70 inhibitors were synthesized based on the HSP70 N-terminal nucleotide that may reduce the viability of resistant cancer cells.18 In addition, HSP70 regulates apoptosis signaling pathways for cell growth and survival, such as Bcl-2 and p53.19 Here, we attempt to clarify the role of HSP70 in HepG2 cells by detecting the effects of specific downregulation of HSP70 protein using siRNA approaches.
Selenium (Se) is an important trace element and necessary for mammalian life; it plays a crucial role in human health and regulates many crucial cellular functions.20,21 Cancer nanotechnology has opened up a new horizon for cancer diagnosis, and Se nanoparticles (SeNPs) have exhibited higher anticancer efficacy and lower side effects compared to inorganic and organic selenocompounds.22 SeNPs with their small size and higher surface-to-volume ratio are attracting increasing attention for their excellent bioavailability.23 As special Se species with application potential as drug carriers, SeNPs have attracted much attention in drug discovery.24 Liu et al25 reported the use of SeNPs as carriers of 5-fluorouracil to enhance anticancer efficacy. SeNPs’ conjugation of transferrin could increase their anticancer activity as reported by Huang et al.26 SeNPs functionalized with polyethylene glycol were designed to overcome drug resistance in hepatocellular carcinoma HepG2 cells by Zheng et al.27 SeNPs were used with surface decoration by Gracilaria lemaneiformis polysaccharide to achieve enhanced anticancer efficacy by Jiang et al.28 SeNPs are superior because they are degradable in vivo and can be used as nutrients for many kinds of normal cells or as antiproliferative agents for many kinds of cancer cells.29 Functionalized SeNPs could be internalized by cancer cells through endocytosis, which induces apoptotic cell death by triggering mitochondria-mediated apoptosis pathways.30,31 In the present study, we demonstrated the rational design and synthesis of PEI-conjugated SeNPs for siRNA delivery to achieve cancer cell apoptosis. HSP70 mRNA is immediately degraded through complementary pairing with HSP70 siRNA in the RNA-induced silencing complex. HepG2 cells are thereby depleted of HSP70, and their protective function and cellular sensitivity to cell death induced by SeNPs are increased. This study may provide an effective rational template for chemotherapy and gene therapy technology.
Materials and methods
Materials
Branched PEI with a molecular weight of 25 kDa, Na2SeO3, coumarin-6, thiazolyl blue tetrazolium bromide (MTT), propidium iodide, bicinchoninic acid kit, 4′,6′-diamidino-2-phenyindole (DAPI), 2′,7′-dichlorofluorescein diacetate, and caspase-3 substrate (Ac-DEVD-AMC) were obtained from Sigma-Aldrich (St Louis, MO, USA). Terminal transferase dUTP nick end labeling (TUNEL) assay kit and Annexin-V-FLUOS staining kit were purchased from Roche Applied Science (Basel, Switzerland). Vitamin C was purchased from Guangzhou chemical reagent factory (Guangzhou, People’s Republic of China). HSP70 siRNA was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). LO2 (normal human liver cell line) was purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, People’s Republic of China). HepG2 cell line was purchased from American Type Culture Collection (ATCC® CCL-136™; Manassas, VA, USA) and incubated in Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, Thermo Fisher Scientific Inc., Waltham, MA, USA) containing antibiotic and fetal bovine serum (Gibco Life Technologies, Grand Island, NY, USA) (10%), 100 units/mL penicillin, and 50 units/mL streptomycin (HyClone, Thermo Fisher Scientific Inc.) in a humidified incubator at 37°C with 5% CO2 atmosphere.
Preparation and characterization of siRNAs with PEI-modified Se nanoparticles (Se@PEI@siRNA)
Se@PEI@siRNA nanoparticles were prepared as follows: Briefly, 2 mL of 50 mM vitamin C solution was added dropwise into 0.25 mL 0.1 M Na2SeO3 under magnetic stirring for half an hour at room temperature at a final volume of 25 mL. The synthesized SeNPs were centrifuged to remove the untreated chemicals. Then, 2 mL of SeNP solution was added to 3.792 mg PEI and 12 μL of 10 μM siRNA. After stirring for 1 hour, the Se@PEI@siRNA complex was purified by centrifugation at 10,000 rpm for 10 minutes and redispersed in Milli-Q water. The morphology, particle zeta potential, and size distribution of SeNPs, Se@PEI, and Se@PEI@siRNA were characterized by transmission electron microscope and Zetasizer Nano ZS particle analyzer (Malvern Instruments Limited, Manchester, UK).
Determination of siRNA complexation by gel retardation assay
The encapsulation degree between siRNA and Se@PEI was assessed by agarose gel electrophoresis.32,33 Se@PEI@siRNA were loaded onto 1% agarose gels with GoldView at 100 V for 30 minutes in Tris/borate/ethylenediaminetetraacetic acid buffer. Retardation of siRNA mobility was visualized by ultraviolet light and gel photography.
In vitro siRNA release
Se@PEI@siRNA were dispersed in pH 7.4 and pH 5.3. The release rate of siRNA was detected by Eppendorf Biophotometer Plus (Hamburg, Germany) (the wavelength of siRNA was 260 nm). In vitro siRNA release of Se@PEI@siRNA was measured as previously reported.34
Cell culture and cell viability assay
The HepG2 cells were incubated in DMEM containing antibiotic and fetal bovine serum (10%), 100 units/mL penicillin, and 50 units/mL streptomycin in a humidified incubator at 37°C with 5% CO2 atmosphere. The cytotoxicity of Se@PEI@siRNA was determined using MTT assay as previously described.35–37 Briefly, HepG2 cells were seeded in 96-well culture plates at a density of 4×104 cells per well at 37°C for 24 hours. Then, the cells were treated with Se@PEI@siRNA at different concentrations for different periods of time. After treatment, 20 μL/well of MTT solution (5 mg/mL in phosphate-buffered saline [PBS]) was added and incubated for another 5 hours. The medium was removed and replaced with 150 μL/well dimethyl sulfoxide to dissolve the formazan crystals. The color intensity of the formazan solution, which reflects the cell viability, was measured at 570 nm using a microplate spectrophotometer (VersaMax Molecular Devices Corporation, CA, USA). This study was conducted in accordance with the Declaration of Helsinki, and with approval from the Ethics Committee of Guangzhou Women and Children’s Medical Center.
Intracellular localization and cellular uptake of Se@PEI@siRNA
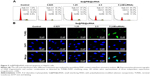
Endocytosis is one of the most important pathways for cellular uptake of extracellular nanoparticles.38–40 To examine the intracellular translocation of the nanoparticles, the localization of Se@PEI@siRNA in HepG2 cells was tracked by simultaneous staining of cell nucleus and lysosome by DAPI and LysoTracker (Thermo Fisher Scientific). Treated cells cultured on a cover glass in 6-well plates were incubated until 70% confluence with different concentrations of coumarin-6-loaded Se@PEI@siRNA for various periods of time. The cells were then washed three times with PBS and examined under a fluorescence microscope. The localization of coumarin-6-labeled Se@PEI@siRNA in HepG2 cells was monitored by DAPI and LysoTracker Red as previously described.41 The mechanisms of cellular uptake of Se@PEI@siRNA in HepG2 cells were investigated by endocytosis inhibitors.42
Flow cytometric analysis
Flow cytometry was used to characterize the effect of Se@PEI@siRNA on cell cycle distribution as previously reported.43 The cells incubated with Se@PEI@siRNA were collected and centrifuged at a speed of 1,500 rpm for 10 minutes. The harvested cells were fixed with 70% ethanol at −20°C overnight followed by propidium iodide staining. The proportion of cells in G0/G1, S, and G2/M phases was represented as a DNA histogram. Apoptotic cells with hypodiploid DNA content were measured by quantifying the sub-G1peak in the cell cycle pattern.
TUNEL assay and DAPI staining
DNA fragmentation was examined with fluorescence staining by the TUNEL apoptosis detection kit according to the manufacturer’s instructions.44 Briefly, HepG2 cells cultured in chamber slides were fixed with 3.7% formaldehyde and then permeabilized with 0.1% Triton X-100 in PBS. The HepG2 cells were labeled with TUNEL reaction mixture for 1 hour. For nuclear staining, the cells were incubated with 1 μg/mL of DAPI for 15 minutes at 37°C. The cells were rinsed with PBS and then observed under a fluorescence microscope (Nikon Eclipse 80i, Tokyo, Japan).
Caspase-3 activity
Caspase activity was determined using a fluorometric method as described previously.45 Harvested cell pellets were suspended in cell lysis buffer and incubated on ice for 1 hour. After centrifugation at 11,000× g for 30 minutes, supernatants were collected and immediately measured for protein concentration and caspase activity. For determination of caspase activity, cell lysates were added in 96-well plates and then incubated with specific caspase-3 substrates (Ac-DEVD-AMC) for 1 hour at 37°C. Caspase-3 activity was determined by fluorescence intensity with the excitation and emission wavelengths set at 380 and 460 nm, respectively.
The efficacy of silencing the HSP70 mRNA by real-time quantitative PCR
The expression of HSP70 was quantitatively detected using real-time quantitative polymerase chain reaction (Q-PCR) technique.46 The following primers were used in Q-PCR experiments. The sequences of primers were as follows: HSP70 primers (forward: 5′-CAGAUUGGAGCAUUUGUGUdTdT-3′, reverse: 5′-ACACAAAUGCUCCAAUCUGdTdT-3′), scrambled siRNA (forward: 5′-AAGAUUCGAGCGUGUGAGUdTdT-3′, reverse: 5′-CCACACAUGCUACAGUCUGdTdT-3′), β-actin primers (forward: 5′-GATATCGCCGCGCTCGTCGTC-3′, and reverse: 5′-TTCTGACCCATGCCCACCATCAC-3′). Scrambled siRNA and Lipofectamine 2000 were used as control to confirm the specificity of the cytotoxic effect of HSP70 knockdown.
Determination of reactive oxygen species generation
Reactive oxygen species (ROS) accumulation induced by Se@PEI@siRNA-treated HepG2 cells was estimated by staining cells with dichlorofluorescein fluorescence assay as previously described.37 Briefly, the cells were harvested and suspended in PBS containing 10 mM of 2′,7′-dichlorofluorescein diacetate. After incubation at 37°C for 30 minutes, the stained cells were collected and resuspended in PBS. Then, the ROS level was determined by measuring the fluorescence intensity using a microplate reader with excitation and emission wavelengths set at 500 and 529 nm, respectively.
Western blotting analysis
The effects of Se@PEI@siRNA on the expression levels of various intracellular proteins in HepG2 cells were detected by Western blotting as previously reported.47,48 HepG2 cells with Se@PEI@siRNA treatment for 24 hours were incubated with lysis buffer to obtain total cellular proteins. The protein concentration was examined by bicinchoninic acid assay. An equal amount of protein was electrophoresed in 12% tricine gels and blocked with 5% non-fat milk in Tris-buffered saline Tween-20 buffer for 1 hour. The membranes were incubated with primary antibodies at 1:1,000 dilutions overnight at 4°C with continuous agitation. Then, the membranes were incubated with secondary antibodies conjugated with horseradish peroxides at 1:1,000 dilutions for 2 hours at room temperature, followed by washing three times with Tris-buffered saline Tween-20 buffer. The bolts were developed with enhanced chemiluminescence reagent using an enhanced chemiluminescence kit that examined the target proteins on the X-ray film.
Statistical analysis
All the data are presented as mean ± standard deviation (SD). Differences between the two groups were evaluated using two-tailed Student’s t-test. One-way analysis of variance was used in multiple-group comparisons. These analyses were carried out by SPSS 13.0 (SPSS Inc., Chicago, IL, USA). A difference of P<0.05 (*) or P<0.01 (**) was considered statistically significant.
Results and discussion
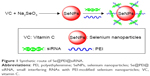
Characterization of Se@PEI@siRNA
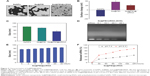
In this study, we demonstrated a simple method to synthesize functionalized SeNPs through self-assembly of functional ligands on the surface of nanoparticles. SeNPs were capped with PEI and siRNA to form more compact and stable globular nanocomposites (Figure 1). The morphology of as-prepared Se@PEI@siRNA was first characterized by transmission electron microscope. Compared with SeNPs and Se@PEI, Se@PEI@siRNA presented uniform and monodisperse spherical particles as shown in Figure 2A. The zeta potential (Figure 2B) of Se@PEI@siRNA with positive charge explains why Se@PEI@siRNA entered more easily into the cell. Regarding size distribution of different nanoparticles were shown in Figure 2C. The binding capacity to HSP70 siRNA was further analyzed by agarose gel electrophoresis as shown in Figure 2D, the binding capability of Se@PEI to HSP70 siRNA was enhanced with the weight ratio. When the weight ratio was over 40:1, almost all the HSP70 siRNAs were restrained with the nanocomposites. As shown in Figure 2E, the result revealed that Se@PEI@siRNA remained stable at least for 30 days. The small size of Se@PEI@siRNA with zeta potential contributed to the highly stability of the nanostructures and the ability to cross the cell membrane. The binding capacity to HSP70 siRNA was further analyzed by agarose gel electrophoresis as shown in Figure 2D; the binding capability of Se@PEI to HSP70 siRNA was enhanced with the weight ratio. When the weight ratio was over 40:1, almost all the HSP70 siRNAs were restrained with the nanocomposites. The siRNA release capacity from Se@PEI@siRNA was detected at pH 7.4 and 5.3 (Figure 2F). The extremely rapid release rate of siRNA from Se@PEI@siRNA at pH 5.3 is due to the acidolysis of nanoparticles. This hydrolysis behavior of siRNA in response to acid pH.
In vitro anticancer activity of Se@PEI@siRNA
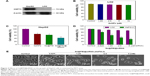
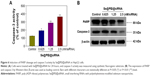
The expression level of HSP70 protein in cancer and normal cells was detected by Western blotting. As shown in Figure 3A, the expression level of HSP70 in HepG2 cells was significantly higher than in LO2 cells, suggesting HSP70-guided selectivity among HepG2 and LO2 cells. The anticancer activity of SeNPs and Se@PEI@siRNA was evaluated by MTT assay, which determines the cells’ survival rate and reflects their growth states. As shown in Figure 3B–D, SeNPs showed nontoxicity as gene delivery systems. Se@PEI@siRNA effectively suppressed HepG2 cells’ growth in a dose-dependent manner, but exhibited low cytotoxicity toward LO2 cells. As shown in Figure 3E, HepG2 cells treated with different concentrations of Se@PEI@siRNA showed cytoplasmic shrinkage, reduction in cells numbers, and loss of cell-to-cell contact. The results suggest that Se@PEI@siRNA effectively inhibited the proliferation of HepG2 cells.
Intracellular localization and cellular uptake of Se@PEI@siRNA
Endocytosis is one of the most important pathways for cellular uptake of extracellular nanoparticles. As shown in Figure 4A, the colocalization of Se@PEI@siRNA and lysosomes was found in HepG2 cells, and increased in a time-dependent manner. Se@PEI@siRNA escaped from lysosomes after 60 minutes and were transported into the cytosol and distributed in the cells after 90 minutes. To further dissect the Se@PEI@siRNA endocytosis mechanism, we used different endocytosis inhibitors to treat the cells before the addition of Se@PEI@siRNA. As shown in Figure 4B, treatments of NaN3 and 2-deoxyglucose markedly inhibited the internalization of Se@PEI@siRNA to 50% of the control. Dynasore and nystatin decreased the Se@PEI@siRNA internalization to 62% and 70% of the control, respectively. Moreover, sucrose induced the uptake of Se@PEI@siRNA to 65%. This result demonstrates that lysosome is the target organelle of Se@PEI@siRNA. Se@PEI@siRNA showed time-dependent cell-penetration for cancer therapy.
Induction of cancer cell apoptosis by Se@PEI@siRNA
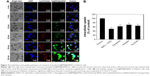
Flow cytometry and TUNEL-DAPI were employed to examine the anticancer mechanisms of Se@PEI@siRNA. As shown in Figure 5A, the sub-G1 apoptotic cell population significantly increased in the DNA histogram in a dose-dependent manner. For instance, Se@PEI@siRNA significantly increased the apoptotic cell population from 1.6% (control) to 55.2% at concentrations of 2.5 nM (siRNA), while no significant change in cell cycle distribution was observed. DNA fragmentation is an important biochemical hallmark of cell apoptosis. As shown in Figure 5B, HepG2 cells exhibited typical apoptotic features with increased concentrations of Se@PEI@siRNA, such as DNA fragmentation and nuclear condensation.
Induction of caspase-mediated poly (ADP-ribose) polymerase (PARP) cleavage by Se@PEI@siRNA
PARP, a DNA repair enzyme, is downstream of caspase family proteins in apoptosis pathways. Caspase-3 has been identified as a key mediator of apoptosis in mammalian cells. Caspase-3 and the subsequent cleavage of PARP were examined by Western blotting to evaluate their involvement and contribution to cell apoptosis. As shown in Figure 6A, the treatment of HepG2 cells with different concentrations of Se@PEI@siRNA significantly increased the activity of caspase-3 in a dose-dependent manner. As shown in Figure 6B, the expression level of caspase-3 and PARP were downregulated with a rise in concentration. The results show that Se@PEI@siRNA significantly enhanced the activation of caspase-3 and the cleavage of downstream effect PARP. Taken together, these results confirm that the synthetic nanosystem inhibits cancer cell growth through inducing cell apoptosis.
Suppression of HSP70 mRNA and protein expression by Se@PEI@siRNA
Inhibition of gene expression has been found to be an effective strategy to suppress cancer growth. In this study, the efficacy of silencing HSP70 gene with Se@PEI@siRNA in HepG2 cells was examined by Q-PCR and Western blotting. As shown in Figure 7A, the level of HSP70 mRNA expression was reduced in the cells exposed to Se@PEI@siRNA. Compared with Se@PEI@scramble siRNA and Lipofectamine 2000@siRNA, the higher silencing efficiency of Se@PEI@siRNA could be due to its positive charge and cancer-targeted delivery. The expression of HSP70 was evaluated by Western blotting. As shown in Figure 7B, Se@PEI@siRNA effectively inhibited the expression of HSP70 in HepG2 cells. Both Q-PCR and Western blotting prove that Se@PEI@siRNA exhibit the ability to silence HSP70 at mRNA and protein levels, which may control the cell fate by regulation of downstream effectors.
Induction of ROS generation by Se@PEI@siRNA
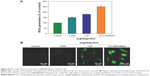
ROS generation has been regarded as a critical regulator of cell apoptosis, especially that induced by anticancer drugs. Mitochondria are the energy factories of cellular activities, but a variety of factors can result in damage to the structure and function of mitochondria and further induce cell apoptosis. ROS were generated from mitochondria that were attacked by Se@PEI@siRNA. Therefore, in this study, ROS generation was determined by DCF fluorescence assay to reveal its role in the action mechanisms of Se@PEI@siRNA. As shown in Figure 8A, the nanosystem increased the intracellular ROS generation in a dose-dependent manner in HepG2 cells. For instance, Se@PEI@siRNA increased the intracellular ROS generation to 180% and 250% at concentrations of 2.5 and 5 nM, respectively. The stronger fluorescent intensity of DCF was also found in HepG2 cells in Figure 8B. These results indicate the involvement of ROS in the anticancer action of Se@PEI@siRNA.
Activation of ROS-mediated signaling pathways by Se@PEI@siRNA
Intracellular ROS overproduction could trigger DNA damage and result in cell apoptosis through the regulation of AKT and p53 signaling pathways. Due to the detection of ROS overproduction in cells exposed to Se@PEI@siRNA, we tried to examine the effects on the ROS-mediated downstream pathways by using Western blotting. As shown in Figure 9A, treatment with Se@PEI@siRNA effectively suppressed the expression levels of total and phosphorylated AKT in HepG2 cells. Similarly, for the p53 signaling pathway, Se@PEI@siRNA significantly increased the expression levels of p-p53 protein and total p53 (Figure 9B). Taken together, these results support the fact that the nanosystem induces cancer cell apoptosis by regulation of ROS-mediated AKT and p53 signaling pathways (Figure 9C).
Conclusion
In summary, PEI-modified SeNPs loaded with HSP70 siRNA have been successfully fabricated in the present study. As the expression of HSP70 was diminished, the cell protective function was also removed, and cancer cells became more sensitive to Se@PEI@siRNA. Se@PEI@siRNA exhibited enhanced cytotoxic effects on human cancer cells through the silencing of HSP70 expression. The gene-silencing efficiency of Se@PEI@siRNA was significantly much higher than Lipofectamine 2000@siRNA. The nanosystem entered cancer cells by endocytosis for intracellular delivery. Moreover, the selective delivery of HSP70 siRNA using the nanosystem resulted in a significantly reduced HSP70 mRNA and protein expression in the cancer cells through the regulation of ROS-mediated downstream signaling pathways, including AKT and p53 pathways. Taken together, this study provides a strategy for design of a nanosystem as a carrier of siRNA for cancer therapy.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No: 309726300), China Postdoctoral Science Foundation (2015M582366), and the Guangdong Natural Science Foundation (No: 10151012001000002).
Disclosure
The authors report no conflicts of interest in this work.
References
Lee MY, Park SJ, Park K, Kim KS, Lee H, Hahn SK. Target-specific gene silencing of layer-by-layer assembled gold-cysteamine/siRNA/PEI/HA nanocomplex. ACS Nano. 2011;5(8):6138–6147. | ||
Li T, Shen X, Chen Y, et al. Polyetherimide-grafted Fe3O4@SiO2 nanoparticles as theranostic agents for simultaneous VEGF siRNA delivery and magnetic resonance cell imaging. Int J Nanomedicine. 2015;10:4279–4291. | ||
Godinho BM, Ogier JR, Quinlan A, et al. PEGylated cyclodextrins as novel siRNA nanosystems: correlations between polyethylene glycol length and nanoparticle stability. Int J Pharm. 2014;473(1–2):105–112. | ||
Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014;194:238–256. | ||
Webb JA, Bardhan R. Emerging advances in nanomedicine with engineered gold nanostructures. Nanoscale. 2014;6(5):2502–2530. | ||
Yang H, Li Y, Li T, et al. Multifunctional core/shell nanoparticles cross-linked polyetherimide-folic acid as efficient Notch-1 siRNA carrier for targeted killing of breast cancer. Sci Rep. 2014;4:7072. | ||
Peng XC, Chen J, Cheng T, et al. PLGA modified Fe3O4 nanoclusters for siRNA delivery. Mater Lett. 2012;81:102–104. | ||
Zou S, Scarfo K, Nantz MH, Hecker JG. Lipid-mediated delivery of RNA is more efficient than delivery of DNA in non-dividing cells. Int J Pharm. 2010;389(1–2):232–243. | ||
Li H, Zheng X, Koren V, Vashist YK, Tsui TY. Highly efficient delivery of siRNA to a heart transplant model by a novel cell penetrating peptide-dsRNA binding domain. Int J Pharm. 2014;469(1):206–213. | ||
Li J, Cheng D, Yin T, et al. Copolymer of poly(ethylene glycol) and poly(L-lysine) grafting polyethylenimine through a reducible disulfide linkage for siRNA delivery. Nanoscale. 2014;6(3):1732–1740. | ||
Morry J, Ngamcherdtrakul W, Gu SD, et al. Dermal delivery of HSP47 siRNA with NOX4-modulating mesoporous silica-based nanoparticles for treating fibrosis. Biomaterials. 2015;66:41–52. | ||
Meneksedag-Erol D, Tang T, Uludag H. Probing the effect of miRNA on siRNA-PEI polyplexes. J Phys Chem. 2015;119(17):5475–5486. | ||
Israel LL, Lellouche E, Ostrovsky S, et al. Acute in vivo toxicity mitigation of PEI-coated maghemite nanoparticles using controlled oxidation and surface modifications toward siRNA delivery. ACS App Mater Interfaces. 2015;7(28):15240–15255. | ||
Wang C, Zhang Y, Guo K, et al. Heat shock proteins in hepatocellular carcinoma: Molecular mechanism and therapeutic potential. Int J Cancer. 2016;138(8):1824–1834. | ||
Wu PK, Hong SK, Veeranki S, et al. A mortalin/HSPA9-mediated switch in tumor-suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol Cell Biol. 2013;33(20):4051–4067. | ||
Shevtsov MA, Nikolaev BP, Yakovleva LY, et al. 70-kDa heat shock protein coated magnetic nanocarriers as a nanovaccine for induction of anti-tumor immune response in experimental glioma. J Control Release. 2015;220(Pt A):329–340. | ||
Gehrmann M, Cervello M, Montalto G, et al. Heat shock protein 70 serum levels differ significantly in patients with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Front Immunol. 2014;5:307. | ||
Zeng Y, Cao R, Zhang T, Li S, Zhong W. Design and synthesis of piperidine derivatives as novel human heat shock protein 70 inhibitors for the treatment of drug-resistant tumors. Eur J Med Chem. 2015;97:19–31. | ||
Kong F, Wang H, Guo J, et al. Hsp70 suppresses apoptosis of BRL cells by regulating the expression of Bcl-2, cytochrome C, and caspase 8/3. In Vitro Cell De Biol Anim. 2016;52(5):568–575. | ||
Ahmad MS, Yasser MM, Sholkamy EN, Ali AM, Mehanni MM. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int J Nanomedicine. 2015;10:3389–3401. | ||
Zheng W, Yin T, Chen Q, et al. Co-delivery of Se nanoparticles and pooled SiRNAs for overcoming drug resistance mediated by P-glycoprotein and class III beta-tubulin in drug-resistant breast cancers. Acta Biomater. 2016;31:197–210. | ||
Yu B, Zhang Y, Zheng W, Fan C, Chen T. Positive surface charge enhances selective cellular uptake and anticancer efficacy of selenium nanoparticles. Inorg Chem. 2012;51(16):8956–8963. | ||
Kumar GS, Kulkarni A, Khurana A, Kaur J, Tikoo K. Selenium nanoparticles involve HSP-70 and SIRT1 in preventing the progression of type 1 diabetic nephropathy. Chem Biol Interact. 2014;223C:125–133. | ||
Kong L, Yuan Q, Zhu H, et al. The suppression of prostate LNCaP cancer cells growth by selenium nanoparticles through Akt/Mdm2/AR controlled apoptosis. Biomaterials. 2011;32(27):6515–6522. | ||
Liu W, Li X, Wong YS, et al. Selenium nanoparticles as a carrier of 5-fluorouracil to achieve anticancer synergism. ACS Nano. 2012;6(8):6578–6591. | ||
Huang Y, He L, Liu W, et al. Selective cellular uptake and induction of apoptosis of cancer-targeted selenium nanoparticles. Biomaterials. 2013;34(29):7106–7116. | ||
Zheng S, Li X, Zhang Y, et al. PEG-nanolized ultrasmall selenium nanoparticles overcome drug resistance in hepatocellular carcinoma HepG2 cells through induction of mitochondria dysfunction. Int J Nanomedicine. 2012;7:3939–3949. | ||
Jiang W, Fu Y, Yang F, et al. Gracilaria lemaneiformis polysaccharide as integrin-targeting surface decorator of selenium nanoparticles to achieve enhanced anticancer efficacy. ACS Appl Mater Interfaces. 2014;6(16):13738–13748. | ||
Tran PA, Sarin L, Hurt RH, Webster TJ. Differential effects of nanoselenium doping on healthy and cancerous osteoblasts in coculture on titanium. Int J Nanomedicine. 2010;5:351–358. | ||
Liu T, Zeng L, Jiang W, Fu Y, Zheng W, Chen T. Rational design of cancer-targeted selenium nanoparticles to antagonize multidrug resistance in cancer cells. Nanomedicine. 2015;11(4):947–958. | ||
Feng Y, Su J, Zhao Z, et al. Differential effects of amino acid surface decoration on the anticancer efficacy of selenium nanoparticles. Dalton Trans. 2014;43(4):1854–1861. | ||
Lin G, Zhu W, Yang L, et al. Delivery of siRNA by MRI-visible nanovehicles to overcome drug resistance in MCF-7/ADR human breast cancer cells. Biomaterials. 2014;35(35):9495–9507. | ||
Li Y, Zhang J, Wang B, Shen Y, Ouahab A. Co-delivery of siRNA and hypericin into cancer cells by hyaluronic acid modified PLGA-PEI nanoparticles. Drug Dev Ind Pharm. 2016;42(5):737–746. | ||
Xiao Y, Jaskula-Sztul R, Javadi A, et al. Co-delivery of doxorubicin and siRNA using octreotide-conjugated gold nanorods for targeted neuroendocrine cancer therapy. Nanoscale. 2012;4(22):7185–7193. | ||
Yang F, Tang Q, Zhong X, et al. Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int J Nanomedicine. 2012;7:835–844. | ||
Li Y, Li X, Wong YS, et al. The reversal of cisplatin-induced nephrotoxicity by selenium nanoparticles functionalized with 11-mercapto-1-undecanol by inhibition of ROS-mediated apoptosis. Biomaterials. 2011;32(34):9068–9076. | ||
Li YH, Li XL, Zheng WJ, Fan CD, Zhang YB, Chen TF. Functionalized selenium nanoparticles with nephroprotective activity, the important roles of ROS-mediated signaling pathways. J Mater Chem B. 2013;1(46):6365–6372. | ||
He L, Chen T, You Y, et al. A cancer-targeted nanosystem for delivery of gold(III) complexes: enhanced selectivity and apoptosis-inducing efficacy of a gold(III) porphyrin complex. Angew Chem Int Ed Engl. 2014;53(46):12532–12536. | ||
He LZ, Huang YY, Zhu HL, et al. Cancer-targeted monodisperse mesoporous silica nanoparticles as carrier of ruthenium polypyridyl complexes to enhance theranostic effects. Adv Funct Mater. 2014;24(19):2754–2763. | ||
Fu X, Yang Y, Li X, et al. RGD peptide-conjugated selenium nanoparticles: antiangiogenesis by suppressing VEGF-VEGFR2-ERK/AKT pathway. Nanomedicine. Epub 2016 Mar 4. | ||
Wu HL, Li XL, Liu W, et al. Surface decoration of selenium nanoparticles by mushroom polysaccharides-protein complexes to achieve enhanced cellular uptake and antiproliferative activity. J Mater Chem. 2012;22(19):9602–9610. | ||
Cao W, Zheng W, Chen T. Ruthenium polypyridyl complex inhibits growth and metastasis of breast cancer cells by suppressing FAK signaling with enhancement of TRAIL-induced apoptosis. Sci Rep. 2015;5:9157. | ||
Li XL, Chen T, Wong YS, et al. Involvement of mitochondrial dysfunction in human islet amyloid polypeptide-induced apoptosis in INS-1E pancreatic beta cells: An effect attenuated by phycocyanin. Int J Biochem Cell Biol. 2011;43(4):525–534. | ||
Yu B, Li XL, Zheng WJ, Feng YX, Wong YS, Chen TF. pH-responsive cancer-targeted selenium nanoparticles: a transformable drug carrier with enhanced theranostic effects. J Mater Chem B. 2014;2(33):5409–5418. | ||
Li XL, Xu G, Chen T, et al. Phycocyanin protects INS-1E pancreatic beta cells against human islet amyloid polypeptide-induced apoptosis through attenuating oxidative stress and modulating JNK and p38 mitogen-activated protein kinase pathways. Int J Biochem Cell Biol. 2009;41(7):1526–1535. | ||
Li JM, Wang YY, Zhao MX, et al. Multifunctional QD-based co-delivery of siRNA and doxorubicin to HeLa cells for reversal of multidrug resistance and real-time tracking. Biomaterials. 2012;33(9):2780–2790. | ||
Zhou L, Duan XM, Zeng S, et al. Codelivery of SH-aspirin and curcumin by mPEG-PLGA nanoparticles enhanced antitumor activity by inducing mitochondrial apoptosis. Int J Nanomedicine. 2015;10:5205–5218. | ||
Zhang YB, Li XL, Huang Z, Zheng WJ, Fan CD, Chen TF. Enhancement of cell permeabilization apoptosis-inducing activity of selenium nanoparticles by ATP surface decoration. Nanomedicine. 2013;9(1):74–84. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.