Back to Journals » The Application of Clinical Genetics » Volume 12
MTHFR C677T polymorphism and risk of nonsyndromic cleft lip with or without cleft palate in the Moroccan population
Authors Rafik A, Rachad L, Kone A, Nadifi S
Received 11 November 2018
Accepted for publication 29 January 2019
Published 7 March 2019 Volume 2019:12 Pages 51—54
DOI https://doi.org/10.2147/TACG.S194166
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Amine Rafik,1,2 Laila Rachad,2 Abdou-samad Kone,2 Sellama Nadifi2
1Department of Plastic Surgery, Alfarbi Hospital, Oujda, Morocco; 2Laboratory of Medical Genetics and Molecular Pathology, Faculty of Medicine and Pharmacy Casablanca, University Hassan II, Casablanca, Morocco
Background: Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is one of the most common craniofacial malformations observed. Several studies suggest that the decrease in folate has been associated with a higher risk of NSCL/P. The present study aimed to determine the association of 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism gene with the occurrence of NSCL/P in the Moroccan population.
Methods: MTHFR C677T was genotyped in 52 Moroccan patients and 182 unrelated controls, using a PCR followed by restriction fragment length polymorphism.
Results: The results of the study revealed a genotypic and phenotypic distribution in equilibrium with Hardy–Weinberg’s law (χ2=0.36, P=0.55). The frequency of heterozygous genotype C/T and the T allele in controls and patients were 40.7% vs 15.4% and 26%, respectively.
Conclusion: A low association was found between the C677T polymorphism of the MTHFR gene and a risk for the development of NSCL/P in the Moroccan population (OR =0.24, P=0.0005).
Keywords: MTHFR, cleft lip and palate, C677T polymorphism, PCR, RFLP
Introduction
Cleft lip with or without cleft palate (CL/P) is one the most prevalent orofacial congenital diseases among children.1,2 The prevalence of oral clefts varies among different ethnic groups, the highest in Asian populations, intermediate in Caucasian populations, and its estimated prevalence is 1.23 per 1,000 live births in Morocco.3 Nonsyndromic CL/P (NSCL/P) is polygenic disease affected by both genetic and environmental factors.4 The genomic revolution has led to major changes in our understanding of the underlying genetics of NSCL/P.
Several lines of evidence have demonstrated a significant association between variations in genes related with folate metabolism, and elevated risk of oral clefts.5 Among genes correlated to folate metabolism, the 5,10-methylenetetrahydrofolate reductase (MTHFR) has been shown to be the most frequent one associated with NSCL/P. This gene is located on chromosome 1 at 1p36.3 and translates to an MTHFR enzyme that catalyzes irreversible reduction of MTHFR to 5-methyltetrahydrofolate that produces the methylation of homocysteine amino acid to methionine.6 Defects in this pathway encode a thermolabile enzyme, leading to reduced enzyme activity and decreased plasma concentration of folate.7 Many studies examined the association of MTHFR gene polymorphisms and the risk of NSCL/P, but their data were contradictory.2,8–12 None of the previous investigations have examined Moroccan patients. Therefore, the aim of this study was to assess the relationship between the MTHFR C677T and the development of NSCL/P in a Moroccan population.
Materials and methods
Sample study
This study was conducted in the Department of Pediatric Surgery at University Hospital EL HAROUCHI, in Casablanca (Morocco), on a total of 52 patients with NSCL/P. The study was approved by the Institutional Ethics Committee of the Faculty of Medicine and Pharmacy, Casablanca, Morocco, and it was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from the parents or guardians and/or the participants. All the patients were examined clinically by two surgeons for their individual phenotypic characteristics, and also a questionnaire of risk factor surveillance for congenital malformation in accordance with US Centers for Disease Control (CDC) was completed. Control patients did not have physical or psychiatric diseases, history of congenital malformations, or familial history of orofacial clefts.
Genotyping
From each study subject, a 2 mL blood sample was collected into an EDTA vacutainer. Genomic DNA of the above samples was isolated by standard protocols with phenol-chloroform extraction and ethanol precipitation. Genotyping for MTHFR C677T polymorphism was performed following published PCR followed by restriction fragment length polymorphism (PCR-RFLP) methods.13,14 The polymorphism at position 677 in the MTHFR gene was studied by PCR followed by Hinf I restriction enzyme digestion. The digested PCR products after separation on a 3% agarose gel, demonstrated one band of 198 bp corresponding to the wild-type homozygous (CC), three bands of 198, 175 and 23 bp for the heterozygous (CT), two bands of 175 and 23 bp for the mutated homozygous (TT).
Statistical analyses
Data were analyzed by using SPSS, version 16.0 (Chicago, IL, USA). The results of continuous variables were expressed as means ± SD. MTHFR gene variants under investigation were evaluated for deviation from Hardy–Weinberg equilibrium analyses (HWP) by comparing observed and expected genotype frequencies by means of χ2 test in case and control groups. Odds ratios ORs and confidence intervals (CIs) were calculated. The results were considered significant at P<0.05.
Results
Fifty-two subjects were available for analysis. All patients had a Moroccan background. The dosages of DNA extracted show very different concentrations between 33 ng/µL for sample 42 and 448 ng/µL for sample 4. However, we can see that the majority of our samples have DNA concentrations >100 ng/µL. Since the DNA concentrations and purity of the samples are known, we will be able to easily adapt the volumes to be used for PCR. The Hinf I RFLP analysis for MTHFR polymorphism is illustrated in Figure 1.
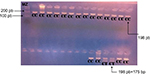  | Figure 1 Results of the digested PCR products by Hinf I enzyme after separation on a 3% agarose gel. CC: homozygosity (198 pb); CT: heterozygosity (198 pb +175 pb); MZ; marker size. |
The frequency of homozygosity for variant C allele is much greater than for variant T allele in C677T. Homozygosity for the C677T MTHFR SNP was detected in 84.61%. The frequency of the C677T heterozygotes was 15.4% (Table 1).
These results show a genotypic and phenotypic distribution of the C677T polymorphism in equilibrium with the Hardy–Weinberg law (χ2=0.36 and P=0.55).
The OR was 0.24 (OR<1), with a CI of 0.1050–0.5367. The P-value for this statistic analysis was very significant, at 0.0005 (P<0.05).
Discussion
NSCL/P is a multifactorial complex that results from interaction between genetic susceptibility and environmental stimulus.15 In this study, we investigated the association of MTHFR C677T polymorphisms with the risk for NSCL/P in a Moroccan population which may help for future diagnosis and early treatment. Fifty-two genotyping was performed by the PCR-RFLP method. The results for the MTHFR variant C677T shows a C/T genotype frequency of 15.4% vs 40.7% in the control population. In addition, the frequencies of the T allele in our study as well as in the control population were, respectively, 0.08% and 26.3%. The control population consisted of 182 healthy patients16 and, by comparing the number of times the T allele was observed in patients and in controls, we conclude that there is an association, but not a strong one, between the variant C677T MTHFR and the occurrence of NSCL/P (OR<1) in the Moroccan population. There are few studies assessing the association of C677T MTHFR polymorphism and risk for NSCL/P, and the results are unclear. Reutter et al17 also found that the MTHFR C677CT polymorphism does not make a major contribution to the occurrence of nonsyndromic cleft lip and palate in the Central European population. In this study, a transmission imbalance test was also performed to define any effect of maternal genotype on the occurrence of splits in neonates. However, the results were not significant and, therefore, showed no evidence of involvement of the maternal genotype in the appearance of this malformation.
Furthermore, a study done by Mills et al18 demonstrated an association between cleft lip palate (CLP) and methylenetetrahydrofolate dehydrogenase 1 in both cases and case mothers in the Ireland population. We also observed similar studies including populations from Norway19 and Iran11 did not support this finding. In fact, Jahanbin et al11 conducted in Iran a study of 45 patients with NSCL/P as well as 43 patient mothers and 101 unrelated controls. This study supports a possible implication of the MTHFR 1298A>C polymorphism, but not the 677C>T variation, in the development of nonsyndromic orofacial clefts. In disagreement with our result, Guo et al20 did not find any significant relationship between the genetic polymorphisms of MTHFR C677T and NSCL/P using the same genotyping techniques. They conclude that variations in the MTHFR gene do not contribute to the development of malformation in the Chinese population.
Conclusion
In conclusion, our data do not suggest that MTHFR C677T is an important risk factor in NSCL/P. However, we are limited by the small sample of patients, which might decrease the statistical power of our study; thus, we recommend further larger studies to determine whether these or additional gene–gene interactions are crucial in determining the probability of carriers of MTHFR alleles presenting an increased or reduced risk of nonsyndromic CL/P.
Acknowledgments
The authors would like to thank Professor M Zemmouri, Head of the Department of Pediatric Surgery, ALHAROUCHI Hospital, the staff at the hospital, and the Genetic Research Center in the Faculty of Medicine and Pharmacy of Casablanca for their kind help in recruiting study subjects.
Disclosure
The authors report no conflicts of interest in this work.
References
Derijcke A, Eerens A, Carels C. The incidence of oral clefts: a review. Br J Oral Maxillofac Surg. 1996;34(6):488–494. | ||
Martinelli M, Scapoli L, Pezzetti F, et al. C677T variant form at the MTHFR gene and CL/P: a risk factor for mothers? Am J Med Genet. 2001;98(4):357–360. | ||
Amine R, Chabbak H, Diouri M, Bahechar N, Chlihi A. Epidemiological profile of clefts lip-palate in Morocco. Int J Oral Maxillofac Surg. 2015;44:e187. | ||
Spritz RA. The genetics and epigenetics of orofacial clefts. Curr Opin Pediatr. 2001;13(6):556–560. | ||
Rafik A, Nadifi S. Updating genetics polymorphisms of non-syndromic clefts lip-palates. Am J Mol Biol. 2018;8(3):178–185. | ||
Goyette P, Pai A, Milos R, et al. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm Genome. 1998;9(8):652–656. | ||
Jacques PF, Bostom AG, Williams RR, et al. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93(1):7–9. | ||
Grunert RR, Braune A, Schnackenberg E, Schloot W, Krause HR. [Genetic differences in enzymes of folic acid metabolism in patients with lip-jaw-palate clefts and their relatives]. Mund Kiefer Gesichtschir. 2002;6(3):131–133. German. | ||
Prescott NJ, Winter RM, Malcolm S. Maternal MTHFR genotype contributes to the risk of non-syndromic cleft lip and palate. J Med Genet. 2002;39(5):368–369. | ||
Pezzetti F, Martinelli M, Scapoli L, et al. Maternal MTHFR variant forms increase the risk in offspring of isolated nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 2004;24(1):104–105. | ||
Jahanbin A, Hasanzadeh N, Abdolhoseinpour F, et al. Analysis of MTHFR Gene C.677C>T and C.1298A>C polymorphisms in iranian patients with non-syndromic cleft lip and palate. Iran J Public Health. 2014;43(6):821–827. | ||
Brandalize APC, Bandinelli E, Borba JB, Félix TM, Roisenberg I, Schüler-Faccini L. Polymorphisms in genes MTHFR, MTR and MTRR are not risk factors for cleft lip/palate in South Brazil. Braz J Med Biol Res. 2007;40:787–791. | ||
Mostowska A, Hozyasz KK, Wojcicki P, Dziegelewska M, Jagodzinski PP. Associations of folate and choline metabolism gene polymorphisms with orofacial clefts. J Med Genet. 2010;47(12):809–815. | ||
Lievers KJ, Boers GH, Verhoef P, et al. A second common variant in the methylenetetrahydrofolate reductase (MTHFR) gene and its relationship to MTHFR enzyme activity, homocysteine, and cardiovascular disease risk. J Mol Med. 2001;79(9):522–528. | ||
Carlson JC, Taub MA, Feingold E, et al. Identifying genetic sources of phenotypic heterogeneity in orofacial clefts by targeted sequencing. Birth Defects Res. 2017;109(13):1030–1038. | ||
Paluku They-They T, Hamzi K, Mazabraud A, Nadifi S. Fréquence Du polymorphisme C677T Du gène de la méthylène tetrahydrofolate réductase (MTHFR) dans les populations arabe et berbère Du Maroc. Antropo. 2009;20:11–17. | ||
Reutter H, Birnbaum S, Lacava AD, et al. Family-based association study of the MTHFR polymorphism C677T in patients with nonsyndromic cleft lip and palate from central Europe. Cleft Palate Craniofac J. 2008;45(3):267–271. | ||
Mills JL, Molloy AM, Parle-McDermott A, et al. Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82(9):636–643. | ||
Al B, Wilcox AJ, Taylor JA, et al. Folate and one-carbon metabolism gene polymorphisms and their associations with oral facial clefts. Am J Med Genet A. 2008;146(4):440–449. | ||
Guo JZ, Song XM, Wang Y, Zhu WL, Li SQ, Li Y. [Relationship between genetic polymorphisms of MTHFR C677T and nonsyndromic cleft lip with or without palate]. Beijing Da Xue Xue Bao Yi Xue Ban. 2009;41(4):432–436. Chinese. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

