Back to Journals » Infection and Drug Resistance » Volume 13
Molecular Relatedness of Salmonella enterica Typhimurium Isolates from Feces and an Infected Surgical Wound
Authors Qin H, Guo Y, Li Y, Zheng R
Received 28 February 2020
Accepted for publication 16 June 2020
Published 6 July 2020 Volume 2020:13 Pages 2139—2144
DOI https://doi.org/10.2147/IDR.S251695
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Haiyan Qin, 1,* Yidan Guo, 2,* Yikun Li, 3, 4 Rui Zheng 3, 4
1Department of Infection Prevention and Control, The First People’s Hospital of Kunming City, Kunming, Yunnan, People’s Republic of China; 2Yunnan Provincial Center of Disease Control and Prevention, Kunming, Yunnan, People’s Republic of China; 3Department of Clinical Laboratory, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, People’s Republic of China; 4Department of Clinical Laboratory, the First People’s Hospital of Yunnan Province, Kunming, Yunnan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Rui Zheng Tel +86 871-63638430
Email [email protected]
Purpose: Salmonella enterica serovar Typhimurium infection is common in foodborne diseases, but its isolation from surgical incisions is rare. Our aim in this study was to trace the transmission source of a surgical incision infected with S. Typhimurium in a Yunnan Province hospital patient and elucidate the underlying molecular mechanisms of antibiotic resistance.
Methods: Primers were designed to amplify the drug-resistance genes using polymerase chain reaction (PCR). Susceptibility to antibiotics was determined using Etest strips. Macrorestriction profiles were analyzed using pulsed-field gel electrophoresis (PFGE) and XbaI. The two isolates were characterized using agglutination tests and multilocus sequence typing (MLST).
Results: MLST analysis revealed that S. Typhimurium isolates SM043 and SM080 belonged to the same genotype, ST34, and PFGE revealed that SM043 and SM080 had high similarity. The isolates were both resistant to third-generation cephalosporins. SM043 harbored the antibiotic resistance genes blaCTX-M-15, blaTEM-1, qnrS-1, qnrB, and acc-3, whereas blaCTX-M-15, blaTEM-1, blaCMY-2, qnrS-1, and acc-3 were detected in SM080.
Conclusion: The surgical incision infection by S. Typhimurium may have been hospital-acquired. Thus, it is critical to strengthen hospital sanitation by addressing hand hygiene and sterilization of the operational environment to avoid outbreaks of nosocomial Salmonella infections.
Keywords: Salmonella, healthcare-associated infection, cephalosporins, antibiotic resistance, nosocomial infection
Corrigendum for this paper has been published
Introduction
Salmonella is a member of the Enterobacteriaceae family, which was originally characterized by the ability of the bacteria to metabolize citrate as a sole carbon source.1 Members of the genus Salmonella cause a well-characterized spectrum of diseases in humans, which range from asymptomatic carriage to fatal typhoid fever. Third-generation cephalosporins are commonly used as first-line antibiotics to treat invasive infections or enteric fevers caused by Salmonella spp., owing to their pharmacodynamic properties and the low prevalence of resistance.2,3 Our previous study found that the drug-resistance rate to third-generation cephalosporins was low in the Yunnan Province.4 Therefore, the mechanism underlying this resistance in Salmonella is still unknown. However, outbreaks or cases of infection caused by Salmonella resistance to extended-spectrum cephalosporins have been reported around the world.5–7 Recently, the growing antibiotic resistance of S. Typhimurium has created new challenges in the control and prevention of potentially fatal infections.8
S. Typhimurium was first isolated in cattle in 1988; the infections it causes are common in foodborne diseases. In China, S. Typhimurium was reported as the most frequently isolated serotype of non-typhoidal Salmonella in foodborne illnesses.9,10 Salmonella infections involving sepsis, liver abscesses, surgical wounds, joint infections, and infected burn wounds have also been described in case reports around the world.9,11 In 2018, two S. Typhimurium isolates resistant to 3rd generation cephalosporins were isolated from a teaching hospital in Yunnan province, China (the name of the hospital has been concealed to maintain anonymity). One of the isolates was from a rare surgical incision secretion. In this study, we aimed to determine the source of the S. Typhimurium to prevent the salmonellosis from spreading or causing an outbreak. The antimicrobial resistance profiles of two S. Typhimurium isolates were analyzed simultaneously to elucidate the molecular type and drug-resistance mechanisms.
Materials and Methods
Bacterial Isolates
Two S. Typhimurium isolates, SM043 and SM080, resistant to third-generation cephalosporins were isolated from clinical specimens in 2018. SM043 was isolated from the feces of a 9-year-old patient with diarrhea and fever on October 30th 2018. On November 5th 2018, SM080 was isolated from an infected incision secretion from a 4-year-old child after surgery owing to a car accident. The two isolates were identified by the VITEK 2 system (bioMérieux, Lyon, France). Serotypes were detected using the White-Kauffmann-Le Minor (WKL) scheme based on serological detection; a diagnostic serum of Salmonella (Statens Serum Institute, Copenhagen, Denmark) was used to determine the serotype.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility was determined by the disk diffusion method and the automated VITEK 2 Compact system with Gram-negative bacteria cards (bioMérieux). The Etest method was employed to verify drug sensitivity to five antibiotics, and the results were interpreted based on CLSI guidelines.12,13 Escherichia coli (ATCC 25,922) was used as the quality control strain for antimicrobial susceptibility testing.
Detection of Drug‑Resistant Genes
Bacterial chromosomal DNA was obtained with a TIANamp Bacterial DNA Kit according to the manufacturer’s instructions (TIANGEN Biotech, Beijing, China). PCR and DNA sequence analysis were performed to confirm the presence of drug-resistance genes. The primers used in this study were described previously.14–20 The β-lactamase genes included Ambler class A (blaCTX-M, blaTEM, blaSHV, blaKPC, and blaGES), class B (blaNDM), class C (blaCMY, blaACT, and blaDHA), and class D (blaOXA-48). Moreover, genes related to quinolone activity included qnrA, qnrB, and qnrS, and the aac gene was also detected. All amplicon sequences were compared with those in the GenBank nucleotide database (www.ncbi.nlm.nih.gov/blast/).
Molecular Typing
The SM043 and SM080 isolates were genotyped using multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE). Seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, thrA) were amplified according to the protocol described on the MLST website (https://pubmlst.org). Genotyping was carried out by referring to the molecular typing method of Salmonella serotype PFGE in PulsenetChina. Salmonella was entrapped with SeaKem Gold Agarose and digested with XbaI. The DNA fragments obtained were separated by PFGE using a Chef Mapper system (Bio-Rad Laboratories, Hercules, CA, USA). Finally, cluster analysis was performed with Bionumerics 7.6 (Applied Maths, Sint-Martens-Latem, Belgium).
Results
Antimicrobial Susceptibility Testing
The drug-resistance profiles of SM043 and SM080 were consistent, showing high resistance to 3rd- and 4th-generation cephalosporins, including ceftriaxone, cefotaxime, and cefepime. Minimum inhibitory concentration values for aminoglycoside and tetracycline were also high. The two isolates were susceptible to β-lactam compound drugs, carbapenems, macrolides, and quinolones. These results are summarized in Table 1.
 |
Table 1 Characteristics of the Two Salmonella Typhimurium Isolates |
Drug-Resistance Genes
A total of 16 drug-resistance genes were amplified, and the two S. Typhimurium isolates carried different genes. The β-lactamase resistance genes blaTEM-1 and blaCTXM-15, quinolone resistance genes qnrB and qnrS-1, and aminoglycoside resistance gene Acc-3 were detected in SM043. The β-lactamase resistance genes blaTEM-1, blaCTXM-15, blaCMY-2, and qnrS-1 were detected in SM080. These results are summarized in Table 1.
Gene Type
MLST analysis showed that SM043 and SM080 were defined as the single sequence type ST34. The two isolates were successfully typed by PFGE and classified into the same PFGE cluster (Figure 1).
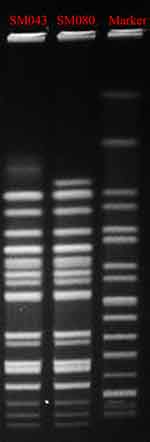 |
Figure 1 Pulsed-field gel electrophoresis patterns of XbaI-digested total DNA from two S. Typhimurium isolates. Abbreviations: MIC, minimum inhibitory concentration; MLST, multilocus sequence typing. |
Discussion
While S. Typhimurium infection is common in foodborne diseases, isolating it from surgical incisions is rare. Two S. Typhimurium isolates were isolated from a hospital in the Yunnan province in 2018, one from an outpatient and one from an inpatient. The first isolate, SM043, was isolated from fecal specimens from a pediatric outpatient with diarrhea on October 30th, 2018. The second, SM080, was isolated from a child who developed a surgical wound infection after emergency trauma surgery on November 5th, 2018. PFGE analysis of the two S. Typhimurium isolates exhibited an identical macrorestriction profile. Further, MLST analysis defined the two S. Typhimurium isolates as the single sequence type ST34. Based on the genotype results of PFGE and MLST analysis, the genetic homology of the two isolates was high.
To reduce the risk of nosocomial transmission, when S. Typhimurium was detected in an incision secretion on November 5th, 2018, necessary disinfection and isolation measures were carried out immediately in the Emergency Trauma Surgery Department. The next morning, environmental swabs from bed linens, stethoscopes, and doorknobs as well as hand swabs from doctors and nurses were collected and tested. However, S. Typhimurium was not isolated from any of the environmental surfaces, likely because of the preventative measures taken. According to field investigation and analysis, we know that both the Emergency Trauma Surgery Department and Pediatric Outpatient Department of the hospital were located in a temporary building owing to construction. Both the children were confirmed to be patients in the temporary hospital building.
Healthcare-associated infections (HAIs) are known to be key causes of morbidity and mortality in hospitalized patients.21 Previous studies have suggested that environmental contamination plays a significant role in HAIs and in the unrecognized transmission of pathogens.22 Cross-transmission of these pathogens can occur via the hands of healthcare workers who become contaminated directly from contact with patients or indirectly by touching contaminated environmental surfaces. S. Typhimurium is highly resistant to stress owing to various survival mechanisms, enabling prolonged survival in harsh environments.23 Unfortunately, in this study, S. Typhimurium could not be isolated from the hospital environment. In an earlier Salmonella outbreak,24 epidemiological investigation revealed that the probable source was leakage of sewage into the water supply system; however, no pathogens were isolated from the water samples. S. Typhimurium may remain in environments for a long time, and several studies have shown that microbial contamination of the ward environment can be a source of nosocomial infection transmission.
Closer investigation and inspection of the temporary hospital structure indicated that the distribution of clinical wards and allocation of resources did not comply with prevention measures established during the transition period. First and foremost, the lack of alcohol hand sanitizer, soap, and tissues in the ward negatively impacted the basic hand hygiene practices of patients and their families as well as the compliance of medical personnel in the wards. In this study, surgical incision infections occurred in children as young as 4 years old who were primarily cared for by family members. Secondly, the building layout is inadequate, especially its toilet configuration. The number of toilets in this transition site, which is an old building, is small, and space is very limited, forcing medical staff, patients, and patient families to share toilets. The utilization rate of these toilets was extremely high. Owing to the lack of hand sanitizers, people could not wash their hands after using the toilets. Thirdly, we also found that the janitorial staff only regularly cleaned and disinfected the wards and ward grounds. The beds, door handles, railings, light switches, window edges, toilet door handles, and other environmental surfaces were not being effectively disinfected, which likely contributed to the spread of bacteria in the environment. The hands of healthcare workers are the main source of pathogen transmission in hospitals. It has been reported that 20–40% of nosocomial infections are caused by microorganisms present in the hospital environment that are transmitted directly or through the hands of healthcare workers.21,25,26 Therefore, in this research, we speculate that S. Typhimurium was present in the hospital environments, serving as potential reservoirs and posing a serious threat to patients. The surgical incision infection caused by S. Typhimurium SM080 was most likely caused by person-to-person transmission. This may have resulted from deteriorating hand hygiene, poor infection control measures, and crowded hospital wards. The nosocomial infection in the Emergency Trauma Surgery Department was finally controlled by the following intervention measures. The most important measure was the uninterrupted provision of hand-washing consumables and improved hand-washing facilities. Second, hand hygiene and nosocomial contact prevention education were provided to patients and staff. Third, the ward custodial team was regularly trained to strengthen cleaning and disinfecting practices. To date, no other nosocomial infections caused by Salmonella have been reported.
In China, cephalosporins and fluoroquinolones are commonly used to treat salmonellosis. Resistance to third-generation cephalosporins is increasing in Salmonella spp. This mainly results from the production of acquired AmpC β-lactamases and extended-spectrum β-lactamases.27 Both SM043 and SM080 harbored at least one AmpC or extended-spectrum β-lactamase or AmpC gene, which made them resistant to 3rd- and 4th-generation cephalosporins. It is important to continuously monitor antimicrobial resistance and genetic status in S. Typhimurium to detect emerging resistance trends. This is the first time that blaTEM-1, blaCTX-M-15, and blaCMY-2 have been detected in S. Typhimurium in the Yunnan province.
Conclusion
In summary, S. Typhimurium spreads not only via the fecal-oral route but also by contact with contaminated hospital environments that serve as potential reservoirs, especially when the strain is resistant to 3rd-generation cephalosporins. Therefore, it is very important to monitor the occurrence of ST34 S. Typhimurium in hospital environments and to take appropriate measures for controlling Salmonella spp. infections.
Abbreviations
PFGE, pulsed-field gel electrophoresis; MLST, multi-locus sequence typing; WKL, White-Kauffmann-Le Minor; HAIs, healthcare-associated infections.
Data Sharing Statement
The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request.
Ethics Statement
The clinical isolates in this study were specifically isolated for this research. Ethical approval was obtained from the Institutional Ethics Committee (The First People’s Hospital of Yunnan Province, Kunming, Yunnan, China). The study protocol was in accordance with the Declaration of Helsinki for Human Research of 1974 (last modified in 2000). The parents of the two patients provided written informed consent before sample collection.
Acknowledgments
All experiments for this study were conducted in the Department of Clinical Laboratory, The First People’s Hospital of Yunnan Province Kunming, Yunnan, China and Department of Clinical Laboratory, The Affiliated Hospital of Kunming University of Science and Technology, Kunming, Yunnan, China.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest.
References
1. Dekker JP, Frank KM. Salmonella, shigella, and yersinia. Clin Lab Med. 2015;35(2):225–246. doi:10.1016/j.cll.2015.02.002
2. Karkey A, Thwaites GE, Baker S. The evolution of antimicrobial resistance in Salmonella Typhi. Curr Opin Gastroenterol. 2018;34(1):25–30. doi:10.1097/MOG.0000000000000406
3. Shane AL, Mody RK, Crump JA, et al. 2017 Infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65(12):e45–e80. doi:10.1093/cid/cix669
4. Yaxian J, Hui Z, Hua N, et al. Antimicrobial resistance surveillance of Salmonella isolates from the First People’s Hospital of Yunnan Province, China. J Infect Dev Ctries. 2015;9(4):333–337. doi:10.3855/jidc.5420
5. Dunne EF, Fey PD, Kludt P, et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA. 2000;284(24):3151–3156. doi:10.1001/jama.284.24.3151
6. Wei Z, Xu X, Yan M, et al. Salmonella typhimurium and Salmonella enteritidis infections in sporadic diarrhea in children: source tracing and resistance to third-generation cephalosporins and ciprofloxacin. Foodborne Pathog Dis. 2019;16(4):244–255. doi:10.1089/fpd.2018.2557
7. Hammami A, Arlet G, Ben Redjeb S, et al. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 beta-lactamase. Eur J Clin Microbiol Infect Dis. 1991;10(8):641–646. doi:10.1007/BF01975816
8. Li W, Li Y, Liu Y, et al. Clonal expansion of biofilm-forming Salmonella typhimurium ST34 with multidrug-resistance phenotype in the southern coastal region of China. Front Microbiol. 2017;8:2090. doi:10.3389/fmicb.2017.02090
9. Liwimbi OM, Komolafe IO. Epidemiology and bacterial colonization of burn injuries in Blantyre. Malawi Med J. 2007;19(1):25–27.
10. Liang Z, Ke B, Deng X, et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009–2012. BMC Infect Dis. 2015;15(1):53. doi:10.1186/s12879-015-0784-4
11. Yan JJ, Chiu CH, Ko WC, Chuang CL, Wu JJ. Ceftriaxone-resistant Salmonella enterica serovar Hadar: evidence for interspecies transfer of blaCMY-2 in a Taiwanese university hospital. J Formos Med Assoc. 2002;101(9):665–668.
12. Humphries RM, Kircher S, Ferrell A, et al. The continued value of disk diffusion for assessing antimicrobial susceptibility in clinical laborato ries: report from the clinical and laboratory standards institute methods development and standardiz ation working group. J Clin Microbiol. 2018;56(8). doi:10.1128/JCM.00437-18.
13. Acharyya S, Sarkar P, Saha DR, Patra A, Ramamurthy T, Bag PK. Intracellular and membrane-damaging activities of methyl gallate isolated from Terminalia chebula against multidrug-resistant Shigella spp. J Med Microbiol. 2015;64(8):901–909. doi:10.1099/jmm.0.000107
14. Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann Lab Med. 2012;32(5):359–361. doi:10.3343/alm.2012.32.5.359
15. P¨¦rez-P¨¦rez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi:10.1128/JCM.40.6.2153-2162.2002
16. Pagani L, Dell’Amico E, Migliavacca R, et al. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae fr om a hospital in northern Italy. J Clin Microbiol. 2003;41(9):4264–4269. doi:10.1128/JCM.41.9.4264-4269.2003
17. Poirel L, H¨¦ritier C, Toln V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. doi:10.1128/AAC.48.1.15-22.2004
18. Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50(8):2872–2874. doi:10.1128/AAC.01647-05
19. Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and cl inical impact. Antimicrob Agents Chemother. 2003;47(8):2385–2392. doi:10.1128/AAC.47.8.2385-2392.2003
20. Wolter DJ, Khalaf N, Robledo IE, et al. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rican Medical Center Hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob Agents Chemother. 2009;53(4):1660–1664. doi:10.1128/AAC.01172-08
21. Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38(5 Suppl 1):S25–33. doi:10.1016/j.ajic.2010.04.196
22. Suleyman G, Alangaden G, Bardossy AC. The role of environmental contamination in the transmission of nosocomial pathogens and healthcare-associated infections. Curr Infect Dis Rep. 2018;20(6):12. doi:10.1007/s11908-018-0620-2
23. de Jonge R, Ritmeester WS, van Leusden FM. Adaptive responses of Salmonella enterica serovar Typhimurium DT104 and other S. Typhimurium strains and Escherichia coli O157 to low pH environments. J Appl Microbiol. 2003;94(4):625–632. doi:10.1046/j.1365-2672.2003.01875.x
24. Kovacic A, Huljev Z, Susic E. Ground water as the source of an outbreak of Salmonella enteritidis. J Epidemiol Glob Health. 2017;7(3):181–184. doi:10.1016/j.jegh.2017.05.001
25. Olmsted RN. Prevention by design: construction and renovation of health care facilities for patient safety and infection prevention. Infect Dis Clin North Am. 2016;30(3):713–728. doi:10.1016/j.idc.2016.04.005
26. McLaws ML. The relationship between hand hygiene and health care-associated infection: it’s complicated. Infect Drug Resist. 2015;8:7–18. doi:10.2147/IDR.S62704
27. Miriagou V, Tassios PT, Legakis NJ, Tzouvelekis LS. Expanded-spectrum cephalosporin resistance in non-typhoid Salmonella. Int J Antimicrob Agents. 2004;23(6):547–555. doi:10.1016/j.ijantimicag.2004.03.006
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
