Back to Journals » Infection and Drug Resistance » Volume 16
Molecular Profile and the Effectiveness of Antimicrobials Drugs Against Staphylococcus aureus and Pseudomonas aeruginosa in the Diagnostic Approaches of Otitis Infection
Authors Almuhayawi MS, Gattan HS , Alruhaili MH, Alharbi MT, Nagshabandi MK , Tarabulsi MK, Almuhayawi SM, Al Jaouni SK, Selim S , Alanazi A, Alruwaili Y, Faried OA, Amin I, Elnosary ME
Received 23 May 2023
Accepted for publication 27 June 2023
Published 5 July 2023 Volume 2023:16 Pages 4397—4408
DOI https://doi.org/10.2147/IDR.S418685
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Mohammed S Almuhayawi,1 Hattan S Gattan,2,3 Mohammed H Alruhaili,1,3 Mohanned Talal Alharbi,4 Mohammed K Nagshabandi,4 Muyassar K Tarabulsi,4 Saad M Almuhayawi,5 Soad K Al Jaouni,6 Samy Selim,7 Awadh Alanazi,7 Yasir Alruwaili,7 Osama Ahmed Faried,8 Islam Amin,9 Mohamed E Elnosary10
1Department of Clinical Microbiology and Immunology, Faculty of Medicine, King AbdulAziz University, Jeddah, 21589, Saudi Arabia; 2Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, 21589, Saudi Arabia; 3Special Infectious Agents Unit, King Fahad Medical Research Center, King AbdulAziz University, Jeddah, 21589, Saudi Arabia; 4Department of Medical Microbiology and Parasitology, Faculty of Medicine, University of Jeddah, Jeddah, 23218, Saudi Arabia; 5Department of Otolaryngology-Head and Neck Surgery, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia; 6Department of Hematology/Oncology, Yousef Abdulatif Jameel Scientific Chair of Prophetic Medicine Application, Faculty of Medicine, King Abdulaziz University, Jeddah, 21589, Saudi Arabia; 7Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Jouf University, Sakaka, 72341, Saudi Arabia; 8Medical Microbiology and Immunology Department, Faculty of Medicine, Beni-Suef University, Beni-Suef, 62513, Egypt; 9Central Laboratory, Ismailia General Hospital, Ismailia, Egypt; 10Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Nasr City, Cairo, 11884, Egypt
Correspondence: Mohammed S Almuhayawi; Samy Selim, Email [email protected]; [email protected]
Background: Otitis externa and otitis media are two types of ear infections that affect people of all ages, although they are more common in newborns and young children. Antibiotic usage, healthcare, and advanced age all play a role in the development of this illness.
Methods: Fifty-eight patients with various kinds of infections of the ears were voluntary patients attending the outpatient clinics of the Prince Mutaib Bin Abdulaziz Hospital in Sakaka, Al Jouf, Saudi Arabia, examined to evaluate the role of bacteria and the likely significance of plasmids in their antibiotic resistance as ear infectious agents.
Results: Staphylococcus aureus and Pseudomonas aeruginosa are the most prevalent bacteria found in ear infections. The greatest number of major bacterial isolates were S. aureus (54%), followed by P. aeruginosa (13%), whereas a smaller number of isolates (3%) were from Streptococcus pyogenes, Bacillus subtilis, and Proteus vulgaris, respectively. Mixed growth was noted in 3.4% of instances. The isolation rate for Gram-positive organisms was 72%, while the rate for Gram-negative species was 28%. All the isolates had DNA greater than 14 kilobases. Hind III analysis of the plasmid DNA extracted from the resistant strains of ear infection demonstrated that antibiotic-resistance plasmids were extensively dispersed. Exotoxin A PCR amplification indicated 396 pb PCR-positive DNA for all identified samples, with the exception of three strains for which no band was observed. Patients in the epidemiological study ranged in number, but all were linked together for the purposes of the study because of their shared epidemiological characteristics.
Conclusion: Vancomycin, linezolid, tigecycline, rifampin, and daptomycin are all antibiotics that have been shown to be effective against S. aureus and P. aeruginosa. Microbiological pattern evaluation and antibiotic sensitivity patterns of the microorganisms providing empirical antibiotics are becoming increasingly crucial to minimize issues and the development of antibiotic-resistant strains.
Keywords: bacterial ear infections, otitis externa, otitis media, Staphylococcus aureus, Pseudomonas aeruginosa, exotoxin A, plasmid, antibiotic resistance, Saudi Arabia
Introduction
Otitis media (OM) is the medical name for an infection of the tympanic membrane or middle ear, and it is characterized by ear pain or discharge.1 Infections in the ear canal can be either acute or chronic.2 An estimated 330 million people throughout the globe suffer from ear infections, with 60% of those individuals losing their hearing.3 Public health issues have arisen in both developing and developed nations because of their high prevalence.4 Adults may become infected with an ear infection as well, although it is more prevalent in children under the age of five. Unnoticed therapy may cause infections to begin in infancy.1 A long-lasting hole may allow bacteria to enter the middle ear. Children are more susceptible to ear infections than adults because their eustachian tubes are not fully mature.5 This permits organisms to enter the nasopharynx more easily. In addition, males had a greater rate of infection than women.6 An ear infection may affect either the inner or outer ear, depending on the source of the infection. Among the three forms are acute otitis media (AOM), otitis media with effusion (OME), and the less common otitis externa, sometimes called swimmer’s ear (SE).7–9 Middle ear tumors, auditory inflammation, and hearing difficulties may all be caused by ear infections.10–12 Ear infections may vary greatly from area to region owing to regional variances in antibiotic prescribing patterns and the occurrence of drug-resistant bacterial strains.13 Ear infections are commonly misdiagnosed due to their symptoms, making it difficult for doctors to pinpoint the specific cause of a patient’s illness.14 Antibiotics may be prescribed regardless of the underlying cause of the illness.15
Bacteria, viruses, or fungi may cause ear infections. Bacteria such as S. aureus, Escherichia coli, P. aeruginosa, Streptococcus pyogenes, Protus mirabilis, Klebsiella spp., and mixed infections are the most common pathogens in ear infections.16,17 Antibiotics are becoming less effective against AOM-inducing pathogens. The conventional antibiotic treatment for ear infections may need to be re-evaluated after this sighting.1,18 Recent antibiotic treatment of acute otitis media, children in daycare centers, wintertime infections, and AOM among children under two years of age are all risk factors for resistant bacteria.19,20 In order to eradicate drug-resistant Streptococcus pneumoniae, a greater dose of amoxicillin (80 mg per kg per day) may be necessary.21 Second-line options include oral cefuroxime or amoxicillin-clavulanate, as well as injectable ceftriaxone.19 The discovery of the R plasmid demonstrates that bacteria have both innate resistance genes and the ability to acquire resistance in order to protect themselves from environmental threats.22 Plasmids and transposons, which allow bacteria to acquire and transfer foreign genes, are regarded as key components in the development of resistance.22–24 Intrinsic resistance in Gram-negative bacteria is linked to the control of outer membrane proteins, such as porins and components of efflux pump systems, on the outer membrane.25,26 Small metabolites, such as antibiotics, are normally controlled by porins, while efflux systems use an energy-dependent method to remove harmful compounds such as antibiotics via specialized efflux pumps.27,28 Thus, antimicrobial resistance is a global problem because of mismanagement or abuse of antimicrobial agents. Experts in these places are also unaware of the antibiotic-resistant tendency in their health areas.
As a result, the objective of this investigation is to inspect the incidence of bacterial agents in ear infections and determine the resistance of these organisms to antibiotics. In addition, this study examined the possible link between antibiotic resistance and the presence of plasmids in the bacterial isolates studied. In the present study, isolates of P. aeruginosa from outpatients were assessed for epidemiological empathy through exhausting genotyping of DNA. Phenotypic and genotypic data were matched to conclude the truthfulness of antibiotic profiles, and molecular analysis outcomes were matched to create a clonal association between medical isolates of P. aeruginosa.
Materials and Methods
Study Design
The study was conducted as a cross-sectional survey from December 2020 to February 2022. Fisher’s formula was used to appraisal the proper sample size. Data and specimens were collected from 58 patients of all sexes and ages who visited Prince Mutaib Bin Abdulaziz Hospital in Sakaka, Al Jouf, Saudi Arabia throughout the research period. Table 1 exhibitions the distribution of ear specimens by gender, age, and location. Specimens that indicated any evidence of contamination were discarded from the examination.
 |
Table 1 Dissemination of Wound Specimens of Patients by Gender, Age and Locations |
Sampling, Isolation, and Identification of Bacteria
All patients hospitalized at Prince Mutaib Bin Abdulaziz Hospital in Sakaka, Al Jouf, Saudi Arabia, suffering from a variety of ear illnesses, volunteered for investigation. Sterilized cotton swabs were used to collect samples from patients with otitis and other illnesses. Separately, 10 mL of alkaline peptone water (5% peptone, pH 8.6) was used to wash the swabs. One- microliter portions of the bacterial suspensions were distributed into Nutrient agar, Blood agar, MacConkey media, Pseudomonas selective medium, and Staphylococcus agar-110. Before enumeration and identification of each plate, the bacteria were incubated face down at 37°C for at least 24–48 hours. Tests on glucose oxidation/fermentation, lactose fermentation, oxygenase, nitrogenase reduction, indole reaction, urease generation and the Voges-Proskauer, arginine and ornithine utilization of pure well-isolated colonies were carried out. The VITEK 2 system (bioMerieux, Marcy l’Etoile, France) assays were used to identify these bacterial species.
Antibiotic Susceptibility Tests of S. aureus and P. aeruginosa
The original nutrient agar stock cultures were used in all studies to circumvent the defeat of antibiotic resistance that might ensue when frequently subculturing bacteria. Five µL sterile broth was used to culture S. aureus, and P. aeruginosa isolates for 18 hours at 37°C. It was then seeded into Müller-Hinton Agar plates with a loopful of 5 µL sterile PBS. After the inoculum had dried, sterile forceps were used to apply antibiotic discs to the infected medium and gently push down to achieve uniform contact. Antibiotic resistance was determined by the width of the inhibitory zones surrounding the antibiotic discs after 48 hours of incubation at 37°C. Oxoid standard antibiotic discs were utilized. After 24 hours of incubation at 37°C, the diameter of clear zones around the antibiotic discs showing bacterial growth inhibition was determined.29 According to the manufacturer’s specifications, bacterial isolates were categorized as “resistant”, “intermediate”, or “sensitive”.30
Plasmid Isolation
Lauryl Broth (LB) tubes and appropriates antibiotics, inoculated with individual bacterial isolates, were incubated in a shaker at 37°C overnight as described by Liu.31
Analysis of Plasmid DNA Restriction
Plasmid DNA was isolated using the density gradient centrifugation method, followed by digestion with the restriction enzyme Hind III (Promega., USA) and electrophoresis on a 0.8% agarose gel to assess the size of the plasmid DNA. Hind III restriction endonuclease purchased from Promega, USA was used to digest the isolated plasmid DNA.
Exotoxins a Gene Detection
PCR was used to detect Ps.aeruginosa from clinical samples by amplifying a 396-bp region of the exotoxin A (ETA) structural gene sequence. PCR amplification of P. aeruginosa (Exotoxin A) specific sequence. The primers used in this experiment and conditions are listed in Table 2.
 |
Table 2 Primers and PCR Condition Used in Detection of Exotoxin a |
Epidemiological Studies
Patients were deliberated to be epidemiological interconnected when they were pickled at the same unit and P. aeruginosa was isolated within five days. A relation number (R) was given to each group of associated patients.
Ethical Considerations
Approval was obtained from the Research Ethics Committee, Jouf University (Ethical Approval No. 3-04-43). All patients provided informed consent, in accordance with the Declaration of Helsinki.
Statistical Analysis
All the data were entered and evaluated by exhausting Statistical Platform for Social Science (SPSS) version 24.
Results
A total of 58 samples were collected comprising of 31% females and 69% males. Most common age group involved was 20–40 years (37.9%) followed by less than 20 years (29.3%) whereas a smaller number of isolates were found in elderly age groups (10.4%) (Table 1). The most predominant bacterial isolates were S. aureus (54%) followed by P. aeruginosa 13%, whereas a smaller number of isolates (3%) were from Streptococcus pyogenes, Bacillus subtilis and Proteus vulgaris respectively (Figure 1). A single growth of organism was found in 56 (96.6%) and mixed growth was observed in 2(3.4%) cases. Occurrence of gram-positive organisms was higher than gram negative cases. With gram positive isolation rate was 72% and for gram negative organisms it was 28%.
 |
Figure 1 Frequency distribution of different bacterial isolates from ear infection. |
Table 3 and Table 4 show the frequency of antibiotic susceptible outline of most frequencies ear infection strains, S. aureus and P. aeruginosa, respectively. The overall resistance rate in S. aureus was resistant to ampicillin (100%), cefoxitin (90.9%), erythromycin (65.9%), and imipenem (43.2%). However, they were all susceptible to vancomycin, linezolid, tigecycline, rifampin, and daptomycin (100%) (Table 3).
 |
Table 3 Frequency of Antibiotic Susceptible Outline of S. Aureus (N= 44) |
The overall resistance rate in P. aeruginosa was 42.9%, with a range of 0% to 100%. There was a high level of resistance rate in the ampicillin, amoxicillin-clavulanate, cefazolin, cefuroxime, ceftriaxone, ertapenem, colistin and tigecycline among P. aeruginosa isolates. However, these isolates were sensitive to ceftazidime, cefepime, piperacillin-tazobactam and all DNA synthesis inhibitors drugs (Table 4).
 |
Table 4 Frequency of Antibiotic Susceptible Outline of P. Aeruginosa (N= 10) |
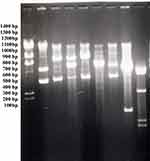
As shown Figure 2 agarose gel profiles of the plasmids after digestion are shown. At least 14 kilobases of plasmid DNA were identified in each one of the isolates. Resistance to ear infection strains can be detected by Hind III digestion of their plasmid DNA . A wide variety of antibacterial-resistant plasmids were discovered.
PCR was used to detect P. aeruginosa from clinical samples by amplifying a 396-bp region of the exotoxin A (ETA) structural gene sequence. We evaluated the specificity of the PCR assay by testing for amplification of the 396-bp DNA from 19 different strains of P. aeruginosa (Figure 3). 84.2% percent of the 19 strains of P. aeruginosa were positive for the ETA gene tested by PCR. Two primers, ETA1 and ETA2 (Table 2), were designed on the basis of the nucleotide sequence of the P. aeruginosa PA103 upstream region of the ETA gene sequence, which is deleted in strain WR5.
Discussion
Every day, OM is a prevalent illness encountered however, it is a non-fatal condition that may cause hearing loss, facial paralysis, and other neurological issues.32 As a result of the low socioeconomic position of persons with limited affordability, the discharging ear is neglected, resulting in insufficient and incorrect treatment. Early detection and adequate treatment are essential to preventing issues.33,34 On the basis of their effectiveness, resistance pattern, cost, and side effects, antibiotics are often prescribed. In order to get the most out of therapy, it is critical to know the antibiotic sensitivity of this disease.35,36 Our findings in our study, 58 samples were obtained comprising 31% females and 69% males. The most common age group involved was 20–40 years (37.9%), followed by 0–20 yrs (29.3%). Related results originated in a study by other investigations who, in their respective studies, found that the common age group involved was 21–30 years, with female predominance (62% and 64.35%) respectively. However, in a study by Wasihun,37 it was seen that the pediatric age group (6–10 yrs) was most commonly involved with male predominance (64.8%).
The most common bacterial isolates were found to be S. aureus (54%) followed by P. aeruginosa 13%, according to our examination. Wasihun et al37 and Agarwal et al38 reported that S. aureus was the most prevalent organism (37.6% and 28.4%, respectively), followed by P. aeruginosa (32.8% and 16.7%) in their investigation; single growth organism was 96.6% in our examination, whereas 3.4% showed mixed growth. Similarly, Agarwal et al38 found monomicrobial bacterial isolates in 80% of patients, whereas only 8% showed mixed growth. Staphylococcus species can be treated effectively with vancomycin, linezolid tigecycline, rifampin, and daptomycin.39 All the cell wall synthesis inhibitors, including ceftazidime, cefepime, piperacillin-tazobactam, and others, had a stronger impact on P. aeruginosa.40 In the current study, the overall resistance rate in S. aureus was resistant to ampicillin (100%), cefoxitin (90.9%), erythromycin (65.9%), and imipenem (43.2%). S. aureus isolates in a similar investigation by Tadesse et al showed the highest level of antimicrobial resistance to ampicillin (100%), clindamycin (63.3%), cephalothin (59.5%), tetracycline (57%), cotrimoxazole and bacitracin (53.2%) and erythromycin (51.9%).41 These results are also in agreement with the reports of Mama et al,42 Soltani et al43 and Okwu et al.44
This investigation found that ear discharge bacterial isolates were MDR (95%). MDR bacteria are on the rise, posing a severe threat to healthcare workers and the general public throughout the world.45 With limited diagnostic resources, insufficient patient education, and widespread non-human antimicrobial use, MDR burdens developing nations more than other places in the world.46 This is especially true in places where there is no formal antimicrobial stewardship policy in place.47 There may be an increased MDR rate in gram negative bacteria owing to the presence of carbapenems enzymes in isolates.48 According to a recent analysis research, are ESL producers, and most of them are classed as MDR because they generally have genes for resistance.49–53
Public health is threatened by the rise of antibiotic-resistant pathogens.54 A broad variety of bacterial species in the environment have antibiotic resistance genes, therefore they are not restricted to the clinic.28 If we want a better understanding of antibiotic resistance, we need to take into account all or most non-producer environmental bacteria’s resistance genes, which may contain determinants that transmit resistance to themselves, as well as those encoding intrinsic resistance mechanisms.55–59 Antibiotic resistance continues to offer severe public health risks for S. aureus. Genes involved in antibiotic resistance in this bacterium include the common ERY resistance genes (ermA, ermB, ermC), and the TET resistance genes (tetL, tetK, tetM, tetO). Transformation (the exchange of nucleic acid), transduction (the insertion into bacteria of a resistance gene mediated by a bacteriophage), conjugation with plasmids or transposons, and mutation are all methods of acquiring resistance. Resistances to high levels of antibiotics have been associated in most instances to the presence of plasmids.60,61 A threat to human health does not arise from the presence of resistance determinants in soil and environmental bacteria; however, when these determinants are mobilized to new hosts and expressed in various contexts, such as plasmids and integrons in pathogenic bacteria, they may become an enormous problem.62–64 Current research has linked antimicrobial-resistant Staphylococci to plasmids (R plasmids) that can facilitate the development of drug inactivated enzymes, such as â-lactamase, and other related functions. Plasmids facilitate the transfer of genetic material, particularly antimicrobial resistance genes, across bacteria of different species and genera.65 Detection of particular strains with possible variations in plasmid content using plasmid profiles is a critical step in epidemiological studies because it is the earliest DNA-based tool for determining serotype-specific reference patterns.66
In the Enterobacteriaceae family, plasmid-mediated antibiotic resistance is well-known. To better understand antibiotic resistance, it has been suggested that examining the plasmid types prevalent in pathogenic bacteria is a vital first step. To examine genetic polymorphism, restriction mapping is one of the most accurate procedures available. Plasmid DNA variation has a distinct restriction nucleotide sequence. It is also possible to identify the original plasmid by looking at the distinctive patterns of DNA bands that appear on agarose after treatment with a single restriction endonuclease. All isolated infection isolates were found to have distinct plasmid patterns. These ten designs were highly diverse. Plasmid variety rises as the number of DNA bands proliferations. Pathogenic bacteria that are isolated from one kind of infection show considerable variance in plasmid. Antibiotic resistance in this disease has been shown through a variety of plasmid restriction maps. Bacterial isolates in this examination were found to be extremely diverse in the DNA of their plasmids.62–64
Endonucleases HindIII, which detect and degrade plasmid DNA molecules at every 5’-AAGCTT-3’, were used to treat the plasmids recovered from pathogenic bacteria that causes ear infection in the present investigation. HindIII is not sensitive to DNA methylation, were chosen in the double digesting procedure for this purpose because of their ability to degrade DNA. Ploidy detection may be accomplished by several approaches, although the majority depend on amplifying certain sequences or genes that are only found in certain kinds of plasmids. Such approaches are used to determine whether a plasmid is present in the bacterium by finding some of the plasmid’s distinctive genes. Because of the large variety of plasmids that identified, restriction mapping was used to identify six-base pair long DNA sequence repeats (restriction sites) in this exertion. Since the HindIII restriction sequences was simple to estimate their distribution here, presenting us with an obvious perspective of plasmid DNA polymorphism.63
There are several kinds of antibiotics now in clinical use that are resistant to plasmid-encoded antimicrobial resistance.22 Cephalosporins, fluoroquinolones, and aminoglycosides are notable examples of these classes of antibiotics.67 In order to facilitate cell-to-cell DNA transfer, many resistance plasmids are conjugative, which encodes the functionalities essential to facilitate their own transfer.68,69 Others may be mobilized with the aid of a cell-resident conjugative plasmid. Genes that allow cells to link prior to DNA transfer are not found in mobilizable plasmids (which are given by the conjugative plasmid), but the genes that encode the activities necessary for the transfer of their own DNA. Thus, whereas conjugative plasmids tend to be much larger than mobilizable resistance plasmids, they are typically less than 10kb in size and encode only a few genes, including the resistance gene(s).70,71 This is because the conjugation functions that allow cell-to-cell coupling, particularly between Gram-negative bacteria, require a substantial amount of DNA (20–30 kb) to encode.72,73 Sex pilus, an exterior filamentous appendage that serves as a grappling hook to unite donor and recipient cells and an envelope-to-envelope contact point when a DNA transfer pore develops to link the cytoplasmic compartments of conjoined cells, mediates this kind of coupling.74 Conjugative plasmids in Gram-positive bacteria tend to be smaller than those in Gram-negative bacteria, suggesting a somewhat different process of cell-to-cell coupling, which necessitates a lower genetic information storage space.75–77 Both the donor and recipient cells get a copy of the plasmid after conjugation, which is a replication event.22
The host range of a conjugative plasmid might be wide or confined, when it comes to the latter, transmission is often confined to and between a few closely related bacterial species.78,79 There are plasmids from Gram-negative bacteria that can transfer between different bacterial species with a broad host range, and these plasmids appear to have no host limitation within the division; and using genetic constructs assembled in the test tube, they can transfer but do not survive in Gram-positive bacteria and unicellular eukaryotic microbes such as yeast.80–82 The resistance plasmid RP1 (also known as RP4 and RK4) was initially discovered in a clinical strain of P. aeruginosa and has since been found in a wide spectrum of bacteria.83,84 Most, if not all, Gram-negative bacteria seem to be able to accept this plasmid. There are other unrelated resistance plasmids with wide host ranges as well.85 Host range is not well understood, but one theory represents the kind of surface receptor required for the plasmid’s specific conjugation machinery in a possible recipient cell.86 This structure must be present in the recipient cell for plasmid transfer to take place. The plasmid’s host range will be restricted if the receptor is only widely distributed. Another scenario is that the recipient cell is unable to sustain the plasmid’s replication, even if the transfer was successful.22 Wide and restricted host range plasmids are not uncommon, although they are not mutually exclusive. Plasmids have been found in the vast majority of bacterial species that have been studied so far, indicating the existence of a sizable reservoir of genetic material that is mobile.28,75 Furthermore, the presence of several plasmids is not at all unusual. In conclusion, the management of this infection will be greatly aided by culture and sensitivity tests, as well as ongoing surveillance to assess antimicrobial resistance and advise antibiotic therapy.
P. aeruginosa’s pathogenicity is influenced by a number of factors; the identification of distinct virulence genes in P. aeruginosa isolates implies that these isolates might be associated with varying degrees of intrinsic virulence and infection propensity. In spite of this, it is important to remember that virulence and other phenotypic features, such as resistance genes, can aid in organism survival, illness dissemination, and severity. Among the several extracellular proteins (LasA and LasB elastases, alkaline protease, protease IV, hemolytic and non-hemolytic phospholipase C, exoenzyme S, and cytotoxin) generated by the opportunistic pathogen P. aeruginosa, the most dangerous is exotoxin A (ETA). ETA is a kind of bacterial toxin that transfers the nicotinamide adenine dinucleotide cation (NAD+adenosine)’s diphosphate (ADP)—ribose moiety to its intended target proteins in eukaryotic cells. Toxins like ETA follow a common A–B structure–function paradigm, where the toxin’s A domain is enzymatically active and the toxin’s B domain binds to a particular receptor on the surface of target cells.87
Conclusion
A single species of microorganism infection, notably S. aureus and P. aeruginosa, was the most prevalent in our investigation. Vancomycin, linezolid, tigecycline, rifampin, and daptomycin were effective against S. aureus. P. aeruginosa is becoming less susceptible to routinely used antimicrobials, such as ampicillin, amoxicillin-clavulanate, cefazolin, cefuroxime, ceftriaxone, ertapenem, colistin and tigecycline. In order to avoid difficulties and the development of antibiotic-resistant strains, it is becoming more important to evaluate the microbiological pattern and the antibiotic sensitivity pattern of the microorganisms to provide empirical antibiotics. In order to prevent the emergence of antibiotic-resistant bacteria and their accompanying consequences, we impulse antimicrobial monitoring and rigorous adherence to antibiotic usage guidelines. Further studies are required to find strains that are resistant to antibiotics utilizing molecular approaches. Most, if not all, kinds of antibiotics currently in clinical use, including resistance to many that are at the forefront of antibiotic therapy, are encoded on plasmids. Plasmids are found in the majority of bacterial species studied to far, revealing a mobile reservoir of transferable genetic material. P. aeruginosa most lethal virulence factor is exotoxin A, which causes intoxication and destroy host cells.
Funding
This research work was funded by Institutional Fund Projects under grant no. (IFPIP: 608-140-1443). The authors gratefully acknowledge technical and financial support provided by the Ministry of Education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Disclosure
The authors report no conflicts of interest in this work
References
1. Danishyar A, Ashurst JV. Acute Otitis Media. Treasure Island (FL): StatPearls Publishing; 2022.
2. Sander RW. Otitis externa: a practical guide to treatment and prevention. AFP. 2001;63:927.
3. Monasta L, Ronfani L, Marchetti F, et al. Burden of disease caused by otitis media: systematic review and global estimates. PLoS One. 2012;7:e36226. doi:10.1371/journal.pone.0036226
4. Deen J, Von Seidlein L, Clemens JD. Issues and challenges of public-health research in developing countries. Manson’s Tropical Infect Dis. 2014;40–48.e1. doi:10.1016/B978-0-7020-5101-2.00006-6
5. Shaikh N, Hoberman A, Kaleida PH, Ploof DL, Paradise JL. Diagnosing otitis media — otoscopy and cerumen removal. N Engl J Med. 2010;362:e62. doi:10.1056/NEJMvcm0904397
6. Ilechukwu GC, Ilechukwu CGA, Ubesie AC, et al. Otitis Media in Children: review Article. OJPed. 2014;4:47–53. doi:10.4236/ojped.2014.41006
7. Klein JO. Otitis Externa, Otitis Media, and Mastoiditis. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Int J Med. 2015:767–773.e1. doi:10.1016/B978-1-4557-4801-3.00062-X
8. Rosenfeld RM, Schwartz SR, Cannon CR, et al. Clinical Practice Guideline: acute Otitis Externa. Otolaryngol Head Neck Surg. 2014;150:S1–S24. doi:10.1177/0194599813517083
9. Lieberthal AS, Carroll AE, Chonmaitree T, et al. The Diagnosis and Management of Acute Otitis Media. Pediatrics. 2013;131:e964–e999. doi:10.1542/peds.2012-3488
10. Byrne AHV, DiMauro E, Frackowiak S, et al. Chapter 97 - The Neurological Vasculitides. In: Neurology and Clinical Neuroscience. Schapira: Philadelphia; 2007:1313–1322.
11. Chronic otitis media (middle ear infection) and hearing loss. Available from: http://www.drrobertoliver.com/what-we-do/sinus-and-ent/chronic-otitis-media-middle-ear-infection-and-hearing-loss.
12. Trojanowska A, Drop A, Trojanowski P, Rosińska-Bogusiewicz K, Klatka J, Bobek-Billewicz B. External and middle ear diseases: radiological diagnosis based on clinical signs and symptoms. Insights Imaging. 2011;3:33–48. doi:10.1007/s13244-011-0126-z
13. Hailu D, Mekonnen D, Derbie A, et al. Pathogenic Bacteria Profile and Antimicrobial Susceptibility Patterns of Ear Infection at Bahir Dar Regional Health Research Laboratory Center, Ethiopia. Springerplus. 2016;5:466. doi:10.1186/s40064-016-2123-7
14. Earwood JS, Rogers TS, Rathjen NA. Ear Pain: diagnosing Common and Uncommon Causes. AFP. 2018;97:20–27.
15. Lin M-F, Lan C-Y. Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World J Clin Cases. 2014;2:787–814. doi:10.12998/wjcc.v2.i12.787
16. Qurban R, Ain QU, Khan JK, et al. Bacterial profile of middle ear infections, coexistence of pseudomonas, proteus species in middle ear infections and their antibiotic sensitivity pattern. Pakistan Postgraduate Med J. 2017;28(4):117.
17. Getaneh A, Ayalew G, Belete D, Jemal M, Biset S. Bacterial Etiologies of Ear Infection and Their Antimicrobial Susceptibility Pattern at the University of Gondar Comprehensive Specialized Hospital, Gondar, Northwest Ethiopia: a Six-Year Retrospective Study. Infect Drug Resist. 2021;14:4313–4322. doi:10.2147/IDR.S332348
18. Kono M, Umar NK, Takeda S, et al. Novel antimicrobial treatment strategy based on drug delivery systems for acute otitis media. Front Pharmacol. 2021;12:640514.
19. Pichichero ME. Acute otitis media: part ii. treatment in an era of increasing antibiotic resistance. Am Fam Physician. 2000;61:2410–2416.
20. Baraibar R. Incidence and risk factors of acute otitis media in children. Clin Microbiol Infection. 1997;3:3S13–3S22. doi:10.1016/S1198-743X(14)64947-8
21. Kaplan SL, Mason EO. Management of infections due to antibiotic-resistant streptococcus pneumoniae. Clin Microbiol Rev. 1998;11:628–644. doi:10.1128/CMR.11.4.628
22. Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153:S347–S357. doi:10.1038/sj.bjp.0707607
23. Sun D, Jeannot K, Xiao Y, Knapp CW. Editorial: horizontal gene transfer mediated bacterial antibiotic resistance. Front Microbiol. 2019;10:10. doi:10.3389/fmicb.2019.00010
24. Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi:10.1146/annurev.biochem.78.082907.145923
25. Amaral L, Martins A, Spengler G, Molnar J. Efflux pumps of gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol. 2014;5:4. doi:10.3389/fphar.2014.00004
26. Fernández L, Hancock REW. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev. 2012;25:661–681. doi:10.1128/CMR.00043-12
27. Li X-Z, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev. 2015;28:337–418. doi:10.1128/CMR.00117-14
28. Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:9. doi:10.3389/fmicb.2018.00009
29. Skov R, Larsen AR, Frimodt-Møller N, Espersen F. Evaluation of different disk diffusion/media combinations for detection of methicillin resistance in Staphylococcus aureus and coagulase-negative staphylococci. APMIS. 2003;111:905–914. doi:10.1034/j.1600-0463.2003.1110909.x
30. Overton N. Oxoid Manual Prelims.
31. Liu D. Handbook of Nucleic Acid Purification. CRC Press; 2009.
32. Vos T, Allen C, Arora M. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 310 Diseases and Injuries, 1990–2015: a Systematic Analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi:10.1016/S0140-6736(16)31678-6
33. Korver AMH, Smith RJH, Van Camp G, et al. Congenital Hearing Loss. Nat Rev Dis Primers. 2017;3:16094. doi:10.1038/nrdp.2016.94
34. Burton MJ, Ramke J, Marques AP, et al. The lancet global health commission on global eye health: vision beyond 2020. Lancet Global Health. 2021;9:e489–e551. doi:10.1016/S2214-109X(20)30488-5
35. Albrecht BA. CHAPTER 61 - Mastitis. In: Samper JC, Pycock JF, McKinnon AO, editors. Current Therapy in Equine Reproduction. W.B. Saunders: Saint Louis; 2007:441–445.
36. Leekha S, Terrell CL, Edson RS. General principles of antimicrobial therapy. Mayo Clinic Proceedings. 2011;86:156. doi:10.4065/mcp.2010.0639
37. Wasihun AG, Zemene Y. Bacterial Profile and Antimicrobial Susceptibility Patterns of Otitis Media in Ayder Teaching and Referral Hospital, Mekelle University, Northern Ethiopia. Springerplus. 2015;4:701. doi:10.1186/s40064-015-1471-z
38. Agrawal A, Kumar D, Goyal A, et al. Microbiological Profile and Their Antimicrobial Sensitivity Pattern in Patients of Otitis Media with Ear Discharge. Indian J Otol. 2013;19:5. doi:10.4103/0971-7749.108149
39. Raad I, Hanna H, Jiang Y, et al. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant staphylococcus bacteremic isolates embedded in biofilm. Antimicrob Agents Chemother. 2007;51:1656–1660. doi:10.1128/AAC.00350-06
40. Pachori P, Gothalwal R, Gandhi P. Emergence of Antibiotic Resistance Pseudomonas Aeruginosa in Intensive Care Unit; a Critical Review. Genes Dis. 2019;6:109–119. doi:10.1016/j.gendis.2019.04.001
41. Tadesse S, Alemayehu H, Tenna A, et al. Antimicrobial re sistance profile of Staphylococcus aureus isolated from patients with infection at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. BMC Pharm Toxicol. 2018;19(1):1–8. doi:10.1186/s40360-018-0210-9
42. Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14. doi:10.1186/1476-0711-13-14
43. Okwu MU, Mitsan OO, Okeke OP. Prevalence and antimicrobial susceptibility profiles of community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) isolates among healthy individuals in Okada, South-South, Nigeria. Pharm Biol Chem Sci J. 2014;1(1):1–9.
44. Soltani R, Khalili H, Rasoolinejad M. Antimicrobial Susceptibility Pattern of Staphylococcus aureus Strains Isolated from Hospitalized Patients in Tehran, Iran. Iranian J Pharm Sci. 2010;6(2):125–132.
45. van Duin D, Paterson D. Multidrug resistant bacteria in the community: trends and lessons learned. Infect Dis Clin North Am. 2016;30:377–390. doi:10.1016/j.idc.2016.02.004
46. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109:309–318. doi:10.1179/2047773215Y.0000000030
47. Doron S, Davidson LE. Antimicrobial Stewardship. Mayo Clin Proc. 2011;86:1113–1123. doi:10.4065/mcp.2011.0358
48. Meletis G. Carbapenem Resistance: overview of the Problem and Future Perspectives. Ther Adv Infect Dis. 2016;3:15–21. doi:10.1177/2049936115621709
49. Tamma PD, Doi Y, Bonomo RA, et al. A Primer on AmpC β-Lactamases: necessary Knowledge for an Increasingly Multidrug-Resistant World. Clin Infect Dis. 2019;69:1446–1455. doi:10.1093/cid/ciz173
50. Fursova NK, Astashkin EI, Knyazeva AI, et al. The Spread of BlaOXA-48 and BlaOXA-244 Carbapenemase Genes among Klebsiella Pneumoniae, Proteus Mirabilis and Enterobacter Spp. Isolated in Moscow, Russia. Ann Clin Microbiol Antimicrob. 2015;14:46. doi:10.1186/s12941-015-0108-y
51. Balasa G, Levengood ES, Battistelli JM, Franklin RB. Diversity of Multidrug-Resistant Bacteria in an Urbanized River: a Case Study of the Potential Risks from Combined Sewage Overflows. Water. 2021;13:2122. doi:10.3390/w13152122
52. Angeli P, Bernardi M, Villanueva C, et al. EASL clinical practice guidelines for the management of patients with decompensated cirrhosis. J Hepatol. 2018;69:406–460. doi:10.1016/j.jhep.2018.03.024
53. Tyne DV, Park DJ, Schaffner SF, et al. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in plasmodium falciparum. PLoS Genet. 2011;7:e1001383. doi:10.1371/journal.pgen.1001383
54. Ventola CL. The antibiotic resistance crisis. P T. 2015;40:277–283.
55. Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proce Royal Society B. 2009;276:2521–2530. doi:10.1098/rspb.2009.0320
56. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance | FEMS Microbiology Reviews | oxford Academic Available from: https://academic.oup.com/femsre/article/42/1/fux053/4563583.
57. Colomer-Lluch M, Jofre J, Muniesa M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS One. 2011;6:e17549. doi:10.1371/journal.pone.0017549
58. Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int. 2016;2016:e2475067. doi:10.1155/2016/2475067
59. Kunhikannan S, Thomas CJ, Franks AE, et al. Environmental Hotspots for Antibiotic Resistance Genes. MicrobiologyOpen. 2021;10:e1197. doi:10.1002/mbo3.1197
60. Pal M, Kerorsa GB, Marami LM, Kandi V. Epidemiology, pathogenicity, animal infections, antibiotic resistance, public health significance, and economic impact of Staphylococcus aureus: a comprehensive review. Am J Public Health Res. 2020;8(1):14–21.
61. Eticha T, Tamire T, Gelgelu TB. Antibiotic susceptibility pattern of coagulase positive Staphylococcus aureus isolated from patients admitted at Tikur Anbessa specialized Hospital, Addis Ababa. Health Sci J. 2022;16(2):1–5.
62. Grenni P. Antimicrobial resistance in rivers: a review of the genes detected and new challenges. Environ Toxicol Chem. 2022;41:687–714. doi:10.1002/etc.5289
63. Gillings MR. Integrons: past, Present, and Future. Microbiol Mol Biol Rev. 2014;78:257–277. doi:10.1128/MMBR.00056-13
64. Palma E, Tilocca B, Roncada P. Antimicrobial resistance in veterinary medicine: an overview. Int J Mol Sci. 2020;21:1914. doi:10.3390/ijms21061914
65. Kot B, Wierzchowska K, Piechota M, Grużewska A. Antimicrobial resistance patterns in methicillin-resistant Staphylococcus aureus from patients hospitalized during 2015–2017 in hospitals in Poland. Med Princ Pract. 2020;29(1):61–68. doi:10.1159/000501788
66. Tamire T, Eticha T, Gelgelu TB. Methicillin-Resistant Staphylococcus aureus: the Magnitude and Risk Factors among Patients Admitted to Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Int J Microbiol. 2021. doi:10.1155/2021/9933926
67. Krause KM, Serio AW, Kane TR, Connolly LE. Aminoglycosides: an Overview. Cold Spring Harb Perspect Med. 2016;6:a027029. doi:10.1101/cshperspect.a027029
68. Frost LS. Conjugation, Bacterial. In: Schaechter M, editor. Encyclopedia of Microbiology (Third Edition). Oxford: Academic Press; 2009:517–531.
69. Zatyka M, Thomas CM. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol Rev. 1998;21:291–319. doi:10.1111/j.1574-6976.1998.tb00355.x
70. Mindlin S, Maslova O, Beletsky A, et al. Ubiquitous conjugative mega-plasmids of Acinetobacter species and their role in horizontal transfer of multi-drug resistance. Front Microbiol. 2021;12:728644.
71. Shearer JES, Wireman J, Hostetler J, et al. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 Genes|Genomes|Genetics. 2011;1:581–591. doi:10.1534/g3.111.000760
72. Bennani S, Nsarellah N, Birouk A, et al. Effective Selection Criteria for Screening Drought Tolerant and High Yielding Bread Wheat Genotypes. Univ J Agr Res. 2016;4:134–142. doi:10.13189/ujar.2016.040404
73. Hawkey PM. Antibiotic Resistance. In: Brenner S, Miller JH, editors. Encyclopedia of Genetics. New York: Academic Press; 2001:74–76.
74. Jin F, Conrad JC, Gibiansky ML, Wong GCL. Bacteria Use Type-IV Pili to Slingshot on Surfaces. Proc Natl Acad Sci U S A. 2011;108:12617–12622. doi:10.1073/pnas.1105073108
75. Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin Microbiol Rev. 2018;31:e00088–17. doi:10.1128/CMR.00088-17
76. Álvarez-Rodríguez I, Arana L, Ugarte-Uribe B, et al. Type IV coupling proteins as potential targets to control the dissemination of antibiotic resistance. Front Mol Biosciences. 2020;7:7. doi:10.3389/fmolb.2020.00007
77. Gössweiner-Mohr N, Arends K, Keller W, Grohmann E. Conjugation in gram-positive bacteria. Microbiology Spectrum. 2014;2. doi:10.13140/2.1.5056.5763
78. Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi:10.1038/nature05775
79. Holmes RK, Jobling MG, Baron S, editors. University of Texas Medical Branch at Galveston: galveston (TX), Genetics. In: In Medical Microbiology; 1996.
80. Yanagiya K, Maejima Y, Nakata H, et al. Novel self-transmissible and broad-host-range plasmids exogenously captured from anaerobic granules or cow manure. Front Microbiol. 2018;9:9.
81. Wright O, Delmans M, Stan G-B, Ellis T. GeneGuard: a Modular Plasmid System Designed for Biosafety. ACS Synth Biol. 2015;4:307–316. doi:10.1021/sb500234s
82. Bartosik AA, Glabski K, Kulinska A, et al. Convenient Broad-Host-Range Unstable Vectors for Studying Stabilization Cassettes in Diverse Bacteria. BMC Microbiol. 2016;16:59. doi:10.1186/s12866-016-0674-y
83. Poole K. Pseudomonas Aeruginosa: resistance to the Max. Front Microbiol. 2011;2:2. doi:10.3389/fmicb.2011.00002
84. Vrancianu CO, Popa LI, Bleotu C, Chifiriuc MC. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front Microbiol. 2020;11:761. doi:10.3389/fmicb.2020.00761
85. Jain A, Srivastava P. Broad host range plasmids. FEMS Microbiol Lett. 2013;348:87–96. doi:10.1111/1574-6968.12241
86. Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi:10.1128/MMBR.65.2.232-260.2001
87. Selim S, Faried OA, Almuhayawi MS, et al. Incidence of vancomycin-resistant Staphylococcus aureus strains among patients with urinary tract infections. Antibiotics. 2022;11:408. doi:10.3390/antibiotics11030408
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.