Back to Journals » Infection and Drug Resistance » Volume 12
Molecular epidemiology and clinical significance of Corynebacterium striatum isolated from clinical specimens
Authors Suh JW, Ju Y, Lee CK, Sohn JW, Kim MJ , Yoon YK
Received 19 August 2018
Accepted for publication 19 November 2018
Published 4 January 2019 Volume 2019:12 Pages 161—171
DOI https://doi.org/10.2147/IDR.S184518
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Jin Woong Suh,1 Yongguk Ju,2 Chang Kyu Lee,3 Jang Wook Sohn,1,2 Min Ja Kim,1,2 Young Kyung Yoon1,2
1Division of Infectious Diseases, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea; 2Institute of Emerging Infectious Diseases, Korea University College of Medicine, Seoul, Republic of Korea; 3Department of Laboratory Medicine, Korea University College of Medicine, Seoul, Republic of Korea
Purpose: This study investigated the clinical epidemiology, antimicrobial susceptibility, and molecular epidemiology of Corynebacterium striatum isolates.
Patients and methods: An observational study was conducted at a university hospital in the Republic of Korea from August to December 2016. All subjects were patients who tested positive for C. striatum clinically. Clinical data were analyzed to evaluate the microbiological and genotypic characteristics of C. striatum strains.
Results: Sixty-seven C. striatum isolates recovered from non-duplicated patients were characterized. Patients were classified into three groups according to the infection type: nosocomial infection (71.6%), health care-associated infection (8.7%), and community-acquired infection (18.8%). The most common clinical specimens were urine (35.8%) and skin abscesses (32.8%). Fifty-two (77.6%) isolates showed multidrug resistance, defined as resistance to ≥3 different antibiotic families. All strains were susceptible to vancomycin and linezolid. Resistance to other antibiotics varied: penicillin (n=65; 97.0%), ampicillin (n=63; 94.0%), cefotaxime (n=64; 95.5%), and levofloxacin (n=61; 91.0%). Phylogenetic analysis identified all 16 S rRNA gene sequences of the 67 isolates as those of C. striatum, where 98%–99% were homologous to C. striatum ATCC 6940. In multilocus sequence typing for internal transcribed spacer region, gyrA, and rpoB sequencing, the most predominant sequence types (STs) were ST2, ST3, ST6, and ST5.
Conclusion: C. striatum isolates may cause opportunistic infections associated with nosocomial infections through horizontal transmission. The presence of multidrug resistance and intra-hospital dissemination implicate C. striatum isolates as a potential target pathogen for infection control and antimicrobial stewardship programs.
Keywords: Corynebacterium striatum, multidrug resistant, multilocus sequence typing, opportunistic infections, nosocomial infections
A corrigendum has been published for this paper read here
Introduction
Most catalase-positive, gram-positive rods, commonly called coryneform or diphtheroid bacteria, have historically been considered simple contaminants that are unlikely to be pathogenic, when isolated from clinical specimens. Of the >80 species of Corynebacterium that have been reported, ~50 species rarely cause infectious diseases in humans.1 Corynebacterium striatum is one of the more commonly isolated coryneform bacteria in clinical microbiology laboratories.
In the last decade, C. striatum has been frequently cultured from various surfaces and medical equipment in hospital settings.2 In clinical settings, C. striatum is increasingly being recognized as a source of opportunistic diseases in immunocompromised patients suffering from malignancies, human immunodeficiency virus infection, or chronic lung diseases, and in patients wearing prosthetic devices. Furthermore, C. striatum is potentially pathogenic in patients with chronic diseases, who are exposed to specific circumstances, such as invasive medical procedures, prolonged use of broad-spectrum antibiotics, or long-term hospitalization.3–5
Occasionally, C. striatum strains are isolated in polymicrobial infections, where their degree of virulence remains largely undetermined. In reality, it is difficult to distinguish between a pathogen causing infection and one causing colonization. Although the clinical significance and prevalence of C. striatum remain unclear, this organism has been responsible for a variety of infections, such as bacteremia, arthritis, osteomyelitis, meningitis, endocarditis, breast abscess, peritonitis, wound infections, and prosthetic joint infections in both immunocompetent and immunocompromised patients.3–6
Infections caused by C. striatum are usually considered to originate endogenously. However, recent studies have documented the possibility of patient-to-patient transmission of C. striatum. The bacterium may cause serious nosocomial infections in intensive care unit patients and spread from patient to patient via physical contact with attending personnel or via the nosocomial environment itself.7–9
Despite early reports of susceptibility to a wide range of antibiotics, multidrug-resistant phenotypes have been recently reported in most C. striatum strains.2,10 Moreover, multidrug-resistant C. striatum may cause nosocomial transmissions resulting in outbreaks in patients with specific risk factors, leading to increased mortality.2,11
There is limited information on the pathogenicity and clinical implications associated with C. striatum strains in a clinical setting. The aim of this study was to investigate the molecular characteristics, clinical significance, and antimicrobial susceptibility of C. striatum, with the goal of increased understanding of its pathogenicity.
Patients and methods
Study design
An observational study was conducted at the Korea University Anam Hospital, a 1,048-bed university hospital in Seoul, Republic of Korea (ROK) from August 2016 to December 2016. All subjects were adult patients (>18 years) whose clinical isolates obtained from diagnostic cultures of clinical specimens tested positive for C. striatum. If multiple C. striatum isolates were recovered from a patient, only the first isolate was included in the study.
This study was performed in compliance with the Declaration of Helsinki. The study protocol was approved by the Institutional Review Board of Korea University Anam Hospital (No. 2018AN0161). As this observational study did not entail any deviations from routine medical practice, the requirement for informed consent was waived.
Clinical data
Clinical data manually extracted from medical records included age, gender, comorbidities, clinical diagnoses, specimen categories, Charlson’s comorbidity index, microbiological data, and treatment outcomes. Determination of the clinical significance of C. striatum isolates was based on clinical findings, such as fever, white blood cell counts, and C-reactive protein, in addition to whether the patient had or had not received antibiotics for C. striatum. Infections were categorized as community acquired, health care associated, or nosocomial, depending on the location that the stains were isolated from. Treatment outcomes were classified on the basis of in-hospital mortality and median length of hospitalization since C. striatum isolation. Additionally, demographic, clinical, and microbiological characteristics between the antibiotic treatment group and the observation group were compared.
C. striatum identification
Processing and incubation of all clinical samples was performed according to routine laboratory protocols. C. striatum isolates were initially identified using the MicroScan WalkAway-96 Plus system (Beckman Coulter, Inc., Brea, CA, USA) and further confirmed by sequencing of the entire 16S rRNA gene.
The C. striatum strains were cultured on blood agar plates for 18 hours at 37°C. The cultured colony was suspended in 200 µL of lysis buffer, incubated overnight at 37°C, and centrifuged for 5 minutes at 3,000 rpm. The pelleted bacteria were suspended in 500 µL of sterile water and boiled for 15 minutes for DNA extraction. C. striatum DNA was extracted using a bacterial AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, ROK). To amplify 16S rRNA, universal primers 16SF27 (5′-AGAGTTTGATCMTGGCTCAG) and 16SR1492 (5′-TACGGYTACCTTGTTACGACTT) were used, as previously described.12 The purified PCR product was sequenced using primer walking with the oligonucleotides using the primers 16SF518 (5′-CCAGCAGCCGCGGTAATAC) and 16SR800 (5′- TACCAGGGTATCTAATCC).13
The amplified sequence was compared with those available in the GenBank database using the BLAST program. 16S rRNA sequences were aligned with Clustal W multisequence alignment program. Phylogenetic trees were constructed using the neighbor-joining genetic distance method and the MEGA 7.0 program package. The Kimura two-parameter model was chosen for all neighbor-joining tree constructions. Reliability of each tree topology was checked with 1,000 bootstrap replications.14 Phylogenetic analysis confirmed that all 16S rRNA gene sequences of the 67 isolates were 98%–99% homologous to C. striatum ATCC 6940 isolates (Figure 1).
Antimicrobial susceptibility
Susceptibility to antibiotics was tested using Streptococcus MicroScan panel (Beckman Coulter, Inc.) and the MicroScan WalkAway-96 Plus system (Beckman Coulter, Inc.), which are considered the gold standard culture media for Corynebacterium as recommended by the Clinical and Laboratory Standards Institute (CLSI). The minimum inhibitory concentration (MIC) breakpoint for Corynebacterium (CLSI document M45-A2) was applied for the analysis of antimicrobial susceptibility.15
Multilocus sequence typing (MLST)
For MLST, the internally transcribed spacer 1 (ITS1) region, as well as gyrA and rpoB were amplified and sequenced for C. striatum strains.2 The primers used are provided in Table S1. PCR amplification and sequence reaction were performed as previously described.16–18
Statistical analyses
Categorical variables are expressed as frequencies and were analyzed using Pearson’s chi-squared test or Fisher’s exact test. Continuous variables are expressed as medians and IQRs. The two-sample t-test or Mann–Whitney U test was used as appropriate to compare continuous variables between groups. Significance was set at P<0.05. SPSS Statistics, version 24.0 (IBM Corporation, Armonk, NY, USA) and SAS 9.4 (SAS Institute Inc., Cary, NC, USA) were used for data analyses.
Results
Patients and clinical characteristics
A total of 67 C. striatum isolates were identified in different patients. Demographic and baseline characteristics of the 67 patients are summarized in Table 1. Twenty (29.9%) patients had received antibiotics for C. striatum isolated from infection sites. Univariate analysis did not indicate significant differences in age, gender, category of infection, and comorbid diseases between the antibiotic treatment and the observation groups (Table 1).
C. striatum was isolated from various types of specimens (Table 2). Urine isolates (35.8%) were the most common, but only 10% of these came from those who received antibiotic therapy. Antibiotics were most commonly used in skin abscess isolates (55.0%; Table 2). Of the 67 specimens, 25 (38.8%) were polymicrobial isolates (Table 2). Among these, Escherichia coli (16.1%), carbapenem-resistant Acinetobacter baumannii (16.1%), and Pseudomonas species (16.1%) were the most common simultaneously isolated strains.
  | Table 2 Microbiological characteristics and treatment outcomes of 67 patients whose clinical specimens tested positive for Corynebacterium striatum |
Antimicrobial susceptibility
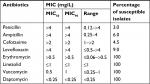
All strains were susceptible to erythromycin, vancomycin, linezolid, and daptomycin (Table 3). However, intermediate to high levels of resistance to penicillin (97.0%), ampicillin (94.0%), cefotaxime (95.5%), and levofloxacin (91.0%) were observed (Table 2). Therefore, the 67 strains may be considered multidrug resistant, defined as resistance to ≥3 classes of antibiotics. There was no significant difference in the frequency of multidrug-resistant isolates between the treatment and observation groups (Table 2).
Based on resistance to penicillin, ampicillin, cefotaxime, levofloxacin, and vancomycin, the 67 C. striatum strains were classified into five patterns, from I to V (Table 4). Among these, pattern I, which was the multidrug-resistant phenotype susceptible to vancomycin, was the predominant in hospitalized patients (82.8%).
Clinical outcomes
Sepsis or septic shock was absent in all cases. Treatment outcomes of the 67 patients are shown in Table 2. Only one (1.5%) patient died during hospitalization. Death was due to a non-infectious cause. Univariate analysis showed no difference between in-hospital mortalities of the treatment and observation groups. The most commonly used therapeutic antibiotics were vancomycin, followed by tigecycline, fluoroquinolones, and piperacillin/tazobactam.
On the other hand, there were no deaths in the 42 patients who had only C. striatum isolated from clinical specimens. There was no difference in the length of hospital stay between the treatment and observation groups (9 [IQR, 0–15] vs 20 [IQR, 4–37] days, P=0.049).
MLST
Four genes (16S rRNA, ITS1, gyrA, and rpoB) were analyzed in all strains studied. 16S rRNA was excluded from the reinforcement analysis of MLST due to its high conservation among all strains analyzed. 16S rRNA was used to confirm the identity of C. striatum strains. ITS1, gyrA, and rpoB were used to discriminate among the strains, using a few nucleotide changes within sequences. Distinct allele sequences were assigned arbitrary allele numbers for each locus (Table S2).
In the ITS1 region, allele 2 was the most abundant (52.2%). For gyrA, allele 1 was predominant (89.6%). For rpoB, allele 2 was the most abundant and was found in 61 strains (91.0%). Twelve sequence types (STs) were identified in these three genes. Of these, the most abundant were ST2 (44.8%), ST3 (22.4%), ST6 (14.9%), and ST5 (4.5%). Among patients with ST2, there were 24 (80.0%) nosocomial infections and 6 (20.0%) community-onset infections. Finally, each ST was grouped and schematized (Figure 2) and analyzed for antibiotic resistance (Table S2). Antibiotic resistance Pattern I occupied an overwhelming number of predominant STs: 28 (93.3%) in ST2, 13 (86.7%) in ST3, and 10 (100%) in ST6.
  | Figure 2 The distribution of STs. Abbreviation: ST, sequence type. |
Discussion
This study analyzed the molecular epidemiology and clinical significance of C. striatum isolates recovered from clinical specimens. The data demonstrate the low virulence of C. striatum, reflecting its role as a colonizing opportunistic pathogen. However, the characteristics of multidrug resistance and horizontal transmission of C. striatum isolates suggest the possibility that it is an emerging nosocomial pathogen, which should be of interest to medical researchers.
C. striatum is reportedly susceptible to β-lactams.19 However, presently, most strains (95.5%) display a multidrug-resistant phenotype, although all strains are susceptible to erythromycin, vancomycin, linezolid, and daptomycin. If C. striatum isolates are, indeed, causative agents of infections, they may require the use of broad-spectrum antibiotics such as vancomycin.20 Antimicrobial resistance is a major global health issue. In ROK, the prevalence of vancomycin-resistant Enterococcus faecium has gradually increased from 22% in 2003 to 31% in 2015.21 Since December 2010, infectious diseases caused by six types of multidrug-resistant bacteria, including vancomycin-resistant enterococci (VRE), were legally designated for surveillance as notifiable infectious microorganisms, with the enactment of the Infectious Disease Control and Prevention Act.22 To control antimicrobial resistance, contamination and infection by C. striatum isolates should be accurately determined to ensure optimal use of antibiotics.
Currently, there are no guidelines for the treatment of infections caused by C. striatum. Optimal antimicrobial therapy is still considered controversial. Our in vitro susceptibility tests demonstrate that vancomycin and linezolid are active against C. striatum, indicating their potential therapeutic value. However, in the absence of approved breakpoints for Corynebacterium species, those antibiotics recommended for Staphylococcus species have been used, except for penicillin and ampicillin, for which thresholds for Listeria species are used.2,23,24
In our study, C. striatum was isolated from various clinical specimens. However, only 20 (29.9%) patients received antibiotic therapy. For several decades, C. striatum has been considered as having limited potential for pathogenicity. The present observations confirmed its low virulence. Similar to C. striatum strains, VRE, which exhibit low virulence and pathogenicity, were initially considered rare opportunistic pathogens. However, in recent years, the prevalence of VRE has increased and they have joined the list of significant pathogens that frequently cause nosocomial infections with limited treatment options.25 Although several factors contribute to virulence, such as host defense mechanisms and the expression of various microorganism traits, a higher rate of multidrug-resistant bacteria may be associated with a higher rate of infection and mortality. Therefore, efforts to contain the spread of multidrug-resistant bacteria should be given prominence by the development of antimicrobial stewardship programs.
Since the first case of C. striatum infection in a patient with chronic lymphocytic leukemia was published in 1980, C. striatum strains have been considered as emerging pathogens in clinical settings.26,27 In recent years, they have been reported as opportunistic pathogens infecting immunocompromised patients with malignancies, COPD, cardiovascular diseases, and diabetes. Other known risk factors include long-term hospitalization, previous use of broad-spectrum antibiotics, and exposure to invasive devices.2,9–11 In our study, 52 patients (77.6%) presented at least one predisposing condition. This suggests that C. striatum may have a significant clinical impact on the patients hospitalized for a long term with comorbidity, or with a history of exposure to broad-spectrum antibiotics, or those who are immunosuppressed, as well as critically ill patients with an implanted indwelling device.
In our study, C. striatum strains mainly caused nosocomial infections, and the four predominant STs in MLST, which accounted for 86.6% of the strains, were ST2, ST3, ST6, and ST5. The 67 strains represent distinct allele combinations (13 STs, considering only three genes: ITS1, gyrA, and rpoB). For gyrA, allele 1 was predominant (89.6%). For rpoB, allele 2 was the most abundant and was found in 61 strains (91.0%). A literature review indicates that these characteristics of C. striatum strains may be a major concern for global health institutions because it is an emergent Gram-positive environmental bacterium which is highly persistent, prevalent, and transmissible person to person and through caregivers, with significant multidrug resistance.2 Furthermore, it has often caused nosocomial outbreaks.2,7–11
Identification of Corynebacterium species is challenging because there are limitations to distinguishing Corynebacterium species based on biochemical profiles using the Api Coryne system. Thus, as in our study, molecular methods, such as gene sequencing, are used as the complementary gold standard for bacterial identification.28,29
Our study had several limitations. Firstly, this study used a single-center design and included a small sample of specimens. Secondly, antibiotic treatment was decided by the treating physician based on his or her interpretation of the microbiological and clinical parameters used in general clinical practice. Finally, the antibiotic susceptibility test did not include gentamicin, as recommended by the CLSI guidelines. Therefore, the possibility that antibiotic resistance of the strains is more extensive cannot be excluded.
Conclusion
C. striatum isolates have been characterized as sources of opportunistic infections associated with nosocomial infections through horizontal transmission. They are only susceptible to a limited number of broad-spectrum antibiotics and can be isolated from clinical specimens due to colonization or contamination. Particularly, characteristics of multidrug resistance and intra-hospital dissemination indicate their potential to be considered as a target pathogen in antimicrobial stewardship programs.
Data sharing statement
Further information and requests for individual deidentified participant data sharing will be fulfilled by contacting the corresponding author YKY ([email protected]) during the 5 years after publication. Clinical and molecular data and research ethics approval information can be shared.
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&B Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI16C1048). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50(10):3152–3158. | ||
Renom F, Gomila M, Garau M, et al. Respiratory infection by Corynebacterium striatum: epidemiological and clinical determinants. New Microbes New Infect. 2014;2(4):106–114. | ||
Martínez-Martínez L, Suárez AI, Rodríguez-Baño J, Bernard K, Muniáin MA. Clinical significance of Corynebacterium striatum isolated from human samples. Clin Microbiol Infect. 1997;3(6):634–639. | ||
Lee PP, Ferguson DA Jr, Sarubbi FA. Corynebacterium striatum: an underappreciated community and nosocomial pathogen. J Infect. 2005;50(4):338–343. | ||
Otsuka Y, Ohkusu K, Kawamura Y, Baba S, Ezaki T, Kimura S. Emergence of multidrug-resistant Corynebacterium striatum as a nosocomial pathogen in long-term hospitalized patients with underlying diseases. Diagn Microbiol Infect Dis. 2006;54(2):109–114. | ||
Funke G, von Graevenitz A, Clarridge JE 3rd, Bernard KA. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10(1):125–159. | ||
Baio PV, Mota HF, Freitas AD, et al. Clonal multidrug-resistant Corynebacterium striatum within a nosocomial environment, Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2013;108(1):23–29. | ||
Brandenburg AH, van Belkum A, van Pelt C, Bruining HA, Mouton JW, Verbrugh HA. Patient-to-patient spread of a single strain of Corynebacterium striatum causing infections in a surgical intensive care unit. J Clin Microbiol. 1996;34(9):2089–2094. | ||
Leonard RB, Nowowiejski DJ, Warren JJ, Finn DJ, Coyle MB. Molecular evidence of person-to-person transmission of a pigmented strain of Corynebacterium striatum in intensive care units. J Clin Microbiol. 1994;32(1):164–169. | ||
Verroken A, Bauraing C, Deplano A, et al. Epidemiological investigation of a nosocomial outbreak of multidrug-resistant Corynebacterium striatum at one Belgian university hospital. Clin Microbiol Infect. 2014;20(1):44–50. | ||
Wang J, Wang Y, Du X, et al. Rapid transmission of multidrug-resistant Corynebacterium striatum among susceptible patients in a tertiary hospital in China. J Infect Dev Ctries. 2016;10(12):1299–1305. | ||
Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester: John Wiley and Sons; 1991:115–175. | ||
Senthilraj R, Prasad GS, Janakiraman K. Sequence-based identification of microbial contaminants in non-parenteral products. Braz J Pharm Sci. 2016;52(2):329–336. | ||
Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42(9):3925–3931. | ||
Clinical and Laboratory Standards Institute. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: CLSI Guideline M45. 3rd ed. Wayne (PA): Clinical and Laboratory Standard Institute; 2016. | ||
Guasp C, Moore ER, Lalucat J, Bennasar A. Utility of internally transcribed 16S-23S rDNA spacer regions for the definition of Pseudomonas stutzeri genomovars and other Pseudomonas species. Int J Syst Evol Microbiol. 2000;50 Pt 4:1629–1639. | ||
Sierra JM, Martinez-Martinez L, Vázquez F, Giralt E, Vila J. Relationship between mutations in the gyrA gene and quinolone resistance in clinical isolates of Corynebacterium striatum and Corynebacterium amycolatum. Antimicrob Agents Chemother. 2005;49(5):1714–1719. | ||
Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42(9):3925–3931. | ||
Soriano F, Zapardiel J, Nieto E. Antimicrobial susceptibilities of Corynebacterium species and other non-spore-forming gram-positive bacilli to 18 antimicrobial agents. Antimicrob Agents Chemother. 1995;39(1):208–214. | ||
Hahn WO, Werth BJ, Butler-Wu SM, Rakita RM. Multidrug-resistant Corynebacterium striatum associated with increased use of parenteral antimicrobial drugs. Emerg Infect Dis. 2016;22(11):1908–1914. | ||
Kim D, Ahn JY, Lee CH, et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) data From 2013 to 2015. Ann Lab Med. 2017;37(3):231–239. | ||
Centers for Disease Control and Prevention. Case definitions for National Notifiable Infectious Diseases; 2011. Available from: http://www.cdc.go.kr/CDC/cms/content/58/12558_view.html. Accessed May 17, 2018. | ||
Weiss K, Laverdière M, Rivest R. Comparison of antimicrobial susceptibilities of Corynebacterium species by broth microdilution and disk diffusion methods. Antimicrob Agents Chemother. 1996;40(4):930–933. | ||
Campanile F, Carretto E, Barbarini D, et al. Clonal multidrug-resistant Corynebacterium striatum strains, Italy. Emerg Infect Dis. 2009;15(1):75–78. | ||
Chiang HY, Perencevich EN, Nair R, et al. Incidence and outcomes associated with infections caused by vancomycin-resistant Enterococci in the United States: systematic literature review and meta-analysis. Infect Control Hosp Epidemiol. 2017;38(2):203–215. | ||
Bowstead TT, Santiago SM. Pleuropulmonary infection due to Corynebacterium striatum. Br J Dis Chest. 1980;74(2):198–200. | ||
Alibi S, Ferjani A, Boukadida J, et al. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci Rep. 2017;7(1):9704. | ||
Gomila M, Renom F, Gallegos M del C, et al. Identification and diversity of multiresistant Corynebacterium striatum clinical isolates by MALDI-TOF mass spectrometry and by a multigene sequencing approach. BMC Microbiol. 2012;12:52. | ||
Alibi S, Ferjani A, Gaillot O, Marzouk M, Courcol R, Boukadida J. Identification of clinically relevant Corynebacterium strains by Api Coryne, MALDI-TOF-mass spectrometry and molecular approaches. Pathol Biol (Paris). 2015;63(4–5):153–157. |
Supplementary materials
  | Table S1 Primers used in the molecular analysis of 67 Corynebacterium striatum strains |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.