Back to Journals » Infection and Drug Resistance » Volume 11
Molecular detection of rifampin, isoniazid, and ofloxacin resistance in Iranian isolates of Mycobacterium tuberculosis by high-resolution melting analysis
Authors Sirous M , Khosravi AD , Tabandeh MR, Salmanzadeh S, Ahmadkhosravi N, Amini S
Received 1 July 2018
Accepted for publication 31 August 2018
Published 18 October 2018 Volume 2018:11 Pages 1819—1829
DOI https://doi.org/10.2147/IDR.S178831
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Mehrandokht Sirous,1,2 Azar Dokht Khosravi,1,2 Mohammad Reza Tabandeh,3 Shokrollah Salmanzadeh,1 Nazanin Ahmadkhosravi,4 Sirus Amini5
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Department of Biochemistry and Molecular Biology, Faculty of Veterinary Medicine, Shahid Chamran University of Ahvaz, Ahvaz, Iran; 4Khuzestan Tuberculosis Regional Reference Laboratory, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 5Tehran Tuberculosis Regional Reference Laboratory, Tehran University of Medical Sciences, Tehran, Iran
Background: The emergence of drug resistance among Mycobacterium tuberculosis (MTB) strains is a serious health concern worldwide. The development of rapid molecular diagnostic methods in recent years has a significant impact on the early detection of resistance to major anti-TB drugs in MTB isolates, which helps in employing appropriate treatment regimen and prevents the spread of drug-resistant strains. This study was designed to evaluate the efficacy of real-time PCR and high-resolution melting (HRM) curve analysis for the determination of resistance to rifampin (RIF), isoniazid (INH), and ofloxacin (OFX) in MTB isolates and to investigate their resistance-related mutations.
Methods: HRM analysis was performed to screen 52 (32 drug-resistant and 20 fully susceptible) MTB clinical isolates for mutations in rpoB, katG, mab-inhA, and gyrA genes. The HRM results were then confirmed by DNA sequencing.
Results: In total, 32 phenotypically resistant isolates, comprising 18 RIF-, 16 INH-, and five OFX- resistant strains, were investigated. HRM analysis successfully identified 15 out of 18 mutations in rpoB, 14 out of 16 mutations in katG and mab-inhA , and four out of five mutations in gyrA conferring resistance to RIF, INH, and OFX, respectively. The obtained sensitivity and specificity, respectively, for HRM in comparison with phenotypic susceptibility testing were found to be 83.3% and 100% for RIF, 87.5% and 100% for INH, and 80% and 100% for OFX. In five resistant strains (12.8%), no mutation was detected by using HRM and DNA sequencing.
Conclusion: HRM assay is a rapid, accurate, and cost-effective method possessing high sensitivity and specificity for the determination of antibiotic resistance among MTB clinical isolates and screening of their associated mutations. This method can generate results in a shorter period of time than taken by the phenotypic susceptibility testing and also allows for timely treatment and prevention of the emergence of possible MDR strains.
Keywords: Mycobacterium tuberculosis, drug resistance, high-resolution melting curve analysis, isoniazid, rifampin, ofloxacin
Introduction
Tuberculosis (TB) still remains a significant global health hazard and is one of the most prevalent life-threatening infections in developing countries. It is the ninth leading cause of death in the world, and according to the latest report of WHO, ~10.4 million people suffered from TB in 2016.1 The emergence of drug-resistant TB, which is associated with increased mortality, is a challenging concern, particularly in developing countries.2
Antibiotic resistance in TB is classified into three main categories: multidrug-resistant TB (MDR-TB) is defined as simultaneous resistance of Mycobacterium tuberculosis (MTB) isolates to rifampin (RIF) and isoniazid (INH). RIF-resistant TB (RR-TB) is the description for MTB isolates that are susceptibe to INH but resistant to RIF. Extensively drug-resistant TB (XDR-TB) is defined as MDR-TB strains with additional resistance to any fluoroquinolone and to at least one of the injectable aminoglycosides.1,3
The management and control of drug-resistant TB, and especially MDR-TB, is difficult due to the poor detection rate, which can be partly attributed to lack of rapid and accurate diagnostic tests. MDR-TB needs long-term treatment with second-line anti-TB drugs which incur high cost and wide toxic effects.2,4 So, rapid detection of antibiotic resistance in TB patients is important and helps in developing strategies aimed to control the spread of drug-resistant disease.
Although the two main effective antibiotics used in the treatment of TB are INH and RIF, drug resistance can develop rapidly as a result of patient non-adherence to the treatment regimen or non-continuous treatment.5 The resistance to RIF is related to rpoB gene, which encodes the β-subunit of bacterial DNA-dependent RNA polymerase, and the significant mutations in rpoB are linked to an 81 bp region, named the rifampicin resistance determinant region (RRDR).2,6 Two mechanisms involving major mutations in katG and inhA genes lead to INH resistance. katG encodes a catalase peroxidase which converts INH to its active form. The amino acid substitution especially at codon 315 of katG, due to a mutation, accounts for 42%–95% of resistance to INH. Another mutation is observed in the promoter region of the inhA gene which encodes the enoyl-ACP- reductase, conferring 6%–34% of INH resistance in MTB strains.7,8
Fluoroquinolones are used as second-line antibiotics for the successful treatment of MDR-TB. Resistance to fluoroquinolones is acquired through mutations within the gyrA and gyrB genes, called the quinolone resistance-determining regions (QRDRs), which encode DNA gyrase in bacteria.9 In a previous study, ofloxacin (OFX) resistance associated with gyrA gene was reported in up to 98% of MTB strains.10
Critical for infection control, the WHO (2017) has recommended widespread use of the drug susceptibility testing (DST), in order to detect the resistant strains of MTB, to prevent the spread of these strains. However, the phenotypic DST is slow, time-consuming, and laborious, so rapid molecular techniques have been currently employed both for the detection of MTB from clinical specimens and detection of their antibiotic resistance.11 These molecular-based assays can be efficiently applied for the rapid identification of genetic mutations and single-nucleotide polymorphisms (SNPs) associated with antibiotic resistance in MTB isolates.12 Molecular sequencing of genes conferring resistance has been recommended as a reference standard for DST in recent years; however, despite the reliability of the tool, it is costly and may not be easily available in all microbiological laboratories.5,13 High-resolution melting (HRM) curve is a simple, rapid, and accurate technique for scanning of mutations, which relies on the sequence variation that leads to changes in the shape of the melting curve.14 By application of this technique, drug resistance among MTB isolates can be demonstrated precisely, which helps in the better management of TB treatment and control of further spread of resistant strains. So, the aims of the current study were to evaluate the efficacy of real-time PCR and HRM curve analysis for determination of mutations in rpoB, katG and mab-inhA, and gyrA genes conferring resistance to RIF, INH, and OFX, respectively, and also in the detection of MDR strains of MTB in the region of study.
Materials and methods
Clinical isolates of MTB
In this study, 487 MTB isolates on the basis of acid-fast staining, growth on Lowenstein Jensen (LJ) medium, and conventional biochemical tests15 were collected from April 2015 to December 2017, from Ahvaz Regional TB Laboratory, Southwest of Iran. The initial proposal of the work was approved by the Institutional Review Board and Ethics Committee of the Ahvaz Jundishapur University of Medical Sciences, Iran, and necessary permission was granted for the work. Apart from this, as part of the Regional Centers’ policy, referred patients were requested to sign the informed consent in case that their samples are used for research purposes apart from routine clinical investigation.
DST
DST was performed for confirmed MTB isolates by proportional method according to the Clinical and Laboratory Standards Institute guidelines.16 This test was done on LJ medium with the following drug concentrations: RIF (40 µg·mL−1), INH (0.2 µg·mL−1), and OFX (4 µg·mL−1) (Sigma Aldrich Co., St Louis, MO, USA). MTB H37Rv (ATCC 27294) was used as the control for the DST. Drug susceptibility was described as no or less than 1% growth on the antibiotic-containing LJ medium as compared with the control medium.
DNA extraction
Genomic DNA was extracted from clinical MTB isolates grown on LJ medium, using DNA extraction QIAamp Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The extracted DNA was diluted tenfold with distilled water and its concentration was measured using a Nanodrop instrument (Thermo Fisher Scientific, Waltham, MA, USA), and was used as template DNA for molecular assays.
PCR amplification and sequence analysis
For the detection of mutations associated with antibiotic resistance, PCR amplification was performed on all phenotypically resistant MTB and randomly selected susceptible isolates. The primers used in the sequence analysis of rpoB, KatG, mab-inhA, and gyrA genes are listed in Table 1.17–20
  | Table 1 Primer sequences used for PCR amplification and sequence analysis |
A reaction mixture of 50 µL containing 5 µL of 10× PCR buffer, 1.5 mM MgCl2, 0.2 mM of deoxynucleotide triphosphate (dNTP), 0.2 µM of each primer, 2.5 U of Taq polymerase, and 5 µL of template DNA (10 ng) was prepared. Amplification was performed on a thermal cycler (Bio-Rad, Hercules, CA, USA) with the following cycling program: initial denaturation at 95°C for 5 minutes followed by 30 cycles of denaturation at 95°C for 40 seconds, annealing at 64°C (rpoB, mab-inhA)/55°C, (katG)/ 68°C, (gyrA) for 1 minute, and extension at 72°C for 40 seconds, with a final extension at 72°C for 10 minutes. The PCR products were sent to Bioneer Co., South Korea, for purification and DNA sequencing (3730xl DNA Analyzers; Thermo Fisher Scientific).
Real-time and HRM curve analysis
The rpoB, katG, mab-inhA, and gyrA genes were amplified by real-time PCR for the determination of mutations associated with RIF, INH, and OFX resistance by using primers listed in Table 2.21,22 The amplification was carried out in a Light Cycler 480 instrument (Roche Diagnostics, Penzberg, Germany) using a Type-it HRM PCR kit (Qiagen). All real-time assays were performed in a mixture containing the following components per reaction: 2 µL of template DNA, 0.2 mM of each primer, 10 µL of 2× HRM PCR master mix containing HotStarTaq DNA Polymerase, HRM PCR buffer, Q-Solution, dNTP mix, and HRM dye with PCR-grade water adjusted to a final volume of 20 µL. The cycling parameters were as follows: 10 minutes at 95°C, followed by 40 cycles of 10 seconds at 95°C, 20 seconds at 64°C, and 20 seconds at 72°C. The products were then heated at 95°C for 1 minute, cooled to 40°C for 1 minute, and heated from 70°C to 95°C at a rate of 25 acquisitions/°C. All real-time reactions were carried out in triplicate. MTB H37Rv DNA and PCR-grade water were used as positive and negative controls, respectively, in every assay.
  | Table 2 Primer sequences used for HRM curve analysis Abbreviation: HRM, high-resolution melting. |
Results
DST
By performing DST, 24 resistant isolates (4.9%) including four MDR and 20 mono-resistant (eleven RIF, six INH, and three OFX) isolates were identified. Additionally, eight more confirmed resistant isolates (two OFX and three INH mono-resistant isolates, and three MDR isolates) provided by Tehran Regional TB Laboratory, Tehran, Iran, were also included in the study. So, in total 32 resistant isolates underwent further molecular investigations.
HRM analysis
In the current study, HRM analysis was employed for the detection of dominant mutations linked to antibiotic resistance in the rpoB, katG, mab-inhA, and gyrA genes in phenotypically resistant MTB clinical isolates. The isolates with mutation were correctly differentiated from the wild-type isolates by differences in their melt curve (Table 3). Different plot curves of HRM analysis for rpoB, katG, mab-inhA, and gyrA genes are also shown in Figure 1 (A–H). For each gene, 20 randomly selected susceptible control isolates were also included. The details of each tested gene are presented in Table 3.
RIF resistance
The 18 phenotypic RIF-resistant isolates were evaluated by HRM analysis for the detection of rpoB gene mutations. There was phenotypic and genotypic concordance (according to HRM curve analysis) in 15 resistant isolates. Four types of curves were generated for these isolates. The most frequent melting curve profile was found in ten isolates which was associated with S531L (TCG→TTG) mutation. The second melting curve profile was obtained for three resistant isolates representing L533P (CTG→CCG) mutation. Other resistant isolates showed two different melting curves which were associated with H526T (CAC→TAC) and L511P (CTG→CCG) mutations (Figure 1A and B). Three phenotypically RIF-resistant isolates did not show any detectable mutation, and these isolates and RIF-susceptible control isolates displayed identical curves by HRM assay.
INH resistance
The 16 phenotypic INH-resistant strains were assessed by HRM analysis for mutations in the katG and mab-inhA genes. There was phenotypic and genotypic concordance (according to HRM curve analysis) in six resistant isolates for katG gene and in eight resistant isolates for mab-inhA gene. The HRM assay successfully detected mutations S315A (AGC→AAC) in all six isolates for katG, and –15 C→T in eight isolates of mab-inhA genes (Figure 1C and D). One isolate had a mutation in mab-inhA on DNA sequencing, but this mutation was not detected by HRM curve analysis. Additionally, in one INH-resistant isolate, no mutation was detected in any of the two target genes on the basis of DNA sequencing and HRM curve analysis. This isolate along with all INH-susceptible control isolates showed unique curves for katG and mab-inhA genes (Figure 1E and F).
OFX resistance
The HRM assay for gyrA gene was carried out using five phenotypic OFX-resistant strains. The phenotypic and HRM analysis methods showed concordance in four OFX-resistant isolates. The HRM curve analysis displayed one mutation at G88C (GGC→TGC) and three at D94G (GAC→GGC) regions of gyrA gene. One resistant isolate did not show any mutation in the gyrA gene according to HRM assay and DNA sequencing. This isolate along with all 20 OFX-susceptible control isolates showed unique curves for gyrA gene (Figure 1G and H).
DNA sequencing
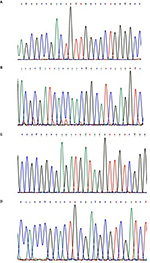
All 52 phenotypic-resistant and -susceptible control MTB isolates underwent sequencing analysis (www.clustal.org) for investigation of mutations in four target genes. DNA sample of MTB H37Rv strain was used as sequence control. Representative examples of the DNA sequencing chromatograms for all target genes are shown in Figure 2. The following results were obtained after sequencing:
The18 phenotypic RIF-resistant strains were sequenced for the rpoB gene mutations. Four mutational profiles were generated for 15 phenotypic-resistant isolates which were confirmed by HRM curve analysis. Three phenotypic-resistant isolates did not show any mutation and displayed wild-type pattern in both DNA sequencing and HRM analysis.
The 16 INH-resistant strains were sequenced for the katG and mab-inhA genes. Six isolates showed mutation pattern of S315A (AGC→AAC) for the katG gene. Eight isolates displayed only one profile of mutation at the –15 C→T of mab-inhA gene. One isolate showed mutation by DNA sequencing at the mab-inhA gene, but did not show any mutation by HRM assay. In one resistant isolate, no mutation was demonstrated in either gene by both sequencing and HRM assay and showed wild-type profile corresponding to 20 INH-susceptible control isolates.
The five phenotypic OFX-resistant strains were sequenced for mutations in the gyrA gene. Four phenotypic-resistant isolates exhibited two different mutations, G88C (GGC→TGC) and D94G (GAC→GGC) for gyrA gene. In other resistant and all susceptible control isolates, no mutation was detected and they showed wild-type pattern.
Statistical analysis
Statistical computations were carried out through the freely available MedCalc statistical software (https://www.medcalc.org/calc/index.php). The HRM analysis assay was compared to the conventional phenotypic results to compute the sensitivity and specificity. The sensitivity and specificity were, respectively, found to be 83.3% and 100% for RIF; 87.5% and 100% for INH; and 80% and 100% for OFX. The comparison of DST and HRM results with sensitivities and specificities for finding out gene mutations are summarized in Table 4.
Nucleotide sequence accession numbers
The nucleotide sequences containing gene mutations in clinical isolates, obtained in the present study, have been deposited in the GeneBank database under the following accession numbers: MH727648, MH727649, MH727650, and MH727651 for rpoB, MH734236 for katG, and MH734237for mab-inhA.
Discussion
Rapid diagnosis of MTB infections and prompt detection of drug resistance, mainly MDR and XDR among the isolates, are essential for the control and management of TB.23,24 The treatment of MDR and XDR-TB is costly and needs vast resources on global basis and makes TB control much more difficult.25 The resistance of MTB to anti-TB drugs is almost exclusively due to the spontaneous mutations in target genes. Molecular methods are rapid tools for the detection of RIF and INH resistance along with resistance to second-line drugs and recognition of their related mutations.26 Among the molecular techniques, real time-PCR coupled with HRM analysis is a rapid and closed-tube method for the screening of DNA mutations in resistant strains.21 HRM allows for the detection of mutations in DNA sequences and sequence variations, which are differentiated by analyzing of the melting curve. This technique could also be used for SNP genotyping and identification of bacterial species.27 Although Xpert-MTB/RIF is recommended by the WHO as a rapid method for the detection of drug-resistant isolates of MTB, this method has limitations, as it only determines resistance to RIF, while HRM can detect resistance to both first- and second-line anti-TB drugs used in the standard treatment regimen.28,29
In the current study, we developed real-time PCR combined with HRM curve analysis for determination of SNP mutations conferring resistance to RIF, INH, and OFX in 32 resistant MTB isolates. The findings showed that HRM assay and DNA sequencing were in concordance for mutation detection in rpoB, katG, and gyrA genes, except for one isolate which showed difference for the mab-inhA gene (Table 3).
The HRM assay successfully determined rpoB gene mutations at codons 531, 533, 526, and 511. Among the 18 RIF-resistant isolates, the majority displayed mutations at codon 531 of 81 bp region (RRDR). According to the literature, this is the region that confers 96% of RIF resistance.6 The second most frequent mutation was found at codon 533, and mutation at codons 526 and 511 were demonstrated in two isolates. Thus, HRM assay could detect the most common rpoB mutations, and based on our results, accounts as preferred method for evaluation of resistance to RIF.
Several genes are responsible for the resistance to INH, including katG, inhA and mab-inhA, kasA, ahpC, and ndh. The major resistance mechanism in INH is katG mutation at codon S315 which is usually associated with high-level resistance. Another less common mechanism is mutation in inhA and mab-inhA genes that causes low-level resistance.30,31 In this study, HRM assay was able to detect mutations at S315A (AGC→AAC) in katG and –15 C→T in mab-inhA. One isolate had mutation in mab-inhA revealed by DNA sequencing, but this mutation was not detected by HRM curve analysis. Additionally, one phenotypic-resistant isolate did not show any mutation by performing either HRM or DNA sequencing. This resistance could be due to other less frequent genes like kasA, ahpC, and ndh, whose roles in INH drug resistance are not well understood.
Clinical isolates of MTB gained fluoroquinolone resistance mainly through mutations in the gyrA gene at QRDR region. Mutations at codons 90, 91, and 94 of gyrA cause high-level resistance to OFX in MTB strains.10,22 Our results demonstrated one mutation at codon 88 and three mutations at codon 94 in the gyrA gene, in four OFX-resistant isolates. One of the phenotypic OFX-resistant isolates did not show any mutation in HRM and DNA sequencing. The resistance in this isolate could be related to mutations in gyrB that we did not examine, or overexpression of efflux pump genes, as other investigators reported.30 Furthermore, in this study, we did not detect any novel mutation by HRM technique or DNA sequencing.
Control of TB in our country requires minimizing the transmission of drug resistance among MTB isolates by employing technologies that allow rapid, cost-effective, and high-throughput detection of resistant isolates.32 In this study, the sensitivity and specificity of real-time HRM assay in comparison to DST were determined as 83.3% and 100% for RIF, and 87.5% and 100% for INH, respectively. The HRM curve analysis has been previously employed by other investigators for the detection of drug resistance among MTB isolates. Our findings were in agreement with these studies and following sensitivities and specificities were observed: 100% and 100% for RIF and 88.8% and 100% for INH by Nagai et al,21 94.4% and 100% for RIF and 95.7% and 97.8% for INH by Chen et al,19 98.7% and 97.5% for RIF and 98.7% and 100% for INH by Galarza et al,33 95% and 100% for RIF and 85.7% and 100% for INH by Haeili et al.34 Although the usefulness of real-time HRM assay is demonstrated in the above-mentioned studies, in a later study conducted in Iran, the investigators concluded that more isolates should be evaluated for the verification of efficacy of HRM assay. Additionally, we carried out determination of gyrA mutation by HRM assay for MDR isolates, and OFX resistance was determined with a sensitivity of 80% and specificity of 100%. It is clear that for the detection of XDR isolates, other second-line drugs, ie, aminoglycosides should be investigated as well.
To determine the frequency of detectable gene mutations related to drug resistance, evaluation of the sensitivity of molecular assays such as HRM curve analysis and DNA sequencing are required.22 In this study, using HRM analysis, the sensitivities of rpoB, katG, and gyrA genes were 100%, which was in agreement with DNA sequencing, except for the sensitivity of 88.89% for mab-inhA. So, HRM assay provides a great potential for characterization of mutations in resistant MTB strains, and is a flexible and low-cost screening technique that can be used in research studies and epidemiological surveys. The significance of HRM assay lies on the fact that it is an effective primary drug resistance indicator among MTB isolates. This enables a fast detection of drug resistance toward first- and second-line anti-TB drugs simultaneously and eliminates the extended period of time (weeks) needed for getting the results by phenotypic DST. However, we failed to detect any mutation in the five phenotypically resistant isolates (12.8%), and hence mutations in other genes with minor frequencies should be considered.
Conclusion
HRM assay is a rapid, accurate, and cost-effective method, which provides high sensitivity and specificity for determination of antibiotic resistance among MTB clinical isolates and screening of their associated mutations. This method would generate results in a shorter period of time than that required by the phenotypic DST method, which allows for timely treatment and prevention of the emergence of possible MDR strains.
Acknowledgments
This work is a part of PhD thesis by Mehrandokht Sirous, which was approved in Infectious and Tropical Diseases Research Center, and was financially supported by a grant (OG-95106) from Research Affairs, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. We are grateful to Dr Mirsaeidi from Department of Medicine, Miller School of Medicine, University of Miami, Florida, USA, for his assistance in manuscript editing.
Disclosure
The authors report no conflicts of interest in this work.
References
WHO. Global tuberculosis report 2017. License: CC BY-NCSA 3.0 IGO. Geneva: World Health Organization; 2017. | ||
Ombura IP, Onyango N, Odera S, Mutua F, Nyagol J. Prevalence of Drug Resistance Mycobacterium Tuberculosis among Patients Seen in Coast Provincial General Hospital, Mombasa, Kenya. PLoS One. 2016;11(10):e0163994. | ||
World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis (2016 update) (WHO/HTM/TB/2016.04). Geneva: WHO; 2016. Available from: http://www.who.int/iris/bitstream/10665/250125/1/9789241549639-eng.pdf. Accessed August 16, 2017. | ||
Dookie N, Rambaran S, Padayatchi N, Mahomed S, Naidoo K. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother. 2018;73(5):1138–1151. | ||
Ramirez MV, Cowart KC, Campbell PJ, et al. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol. 2010;48(11):4003–4009. | ||
Horng Y-T, Jeng W-Y, Chen Y-Y, et al. Molecular Analysis of Codon 548 in the rpoB Gene Involved in Mycobacterium tuberculosis Resistance to Rifampin. Antimicrob Agents Chemother. 2015;59(3):1542–1548. | ||
Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One. 2015;10(3):e0119628. | ||
Bollela VR, Namburete EI, Feliciano CS, Macheque D, Harrison LH, Caminero JA. Detection of katG and inhA mutations to guide isoniazid and ethionamide use for drug-resistant tuberculosis. Int J Tuberc Lung Dis. 2016;20(8):1099–10104. | ||
Maruri F, Sterling TR, Kaiga AW, et al. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother. 2012;67(4):819–831. | ||
Sirgel FA, Warren RM, Streicher EM, Victor TC, van Helden PD, Böttger EC. gyrA mutations and phenotypic susceptibility levels to ofloxacin and moxifloxacin in clinical isolates of Mycobacterium tuberculosis. J Antimicrob Chemother. 2012;67(5):1088–1093. | ||
Almeida da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother. 2011;66(7):1417–1430. | ||
Garberi J, Labrador J, Garberi F, et al. Diagnosis of Mycobacterium tuberculosis using molecular biology technology. Asian Pac J Trop Biomed. 2011;1(2):89–93. | ||
Gilpin C, Korobitsyn A, Weyer K. Current tools available for the diagnosis of drug-resistant tuberculosis. Ther Adv Infect Dis. 2016;3(6):145–151. | ||
Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8(6):597–608. | ||
Tille PM. Bailey & Scott’s Diagnostic Microbiology. 13th ed. St. Louis, MA: Mosby; 2014:484–512. | ||
Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing for Human Mycoplasmas; Approved Guideline. CLSI guideline M43-A. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. | ||
Pozzi G, Meloni M, Iona E, et al. rpoB Mutations in Multidrug-Resistant Strains of Mycobacterium tuberculosis Isolated in Italy. J Clin Microbiol. 1999;37:1197–1199. | ||
Silva MSN, Senna SG, Ribeiro MO, et al. Mutations in katG, inhA, and ahpC Genes of Brazilian Isoniazid-Resistant Isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2003;41(9):4471–4474. | ||
Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL. Rapid Detection of Isoniazid, Rifampin, and Ofloxacin Resistance in Mycobacterium tuberculosis Clinical Isolates Using High-Resolution Melting Analysis. J Clin Microbiol. 2011;49(10):3450–3457. | ||
Fletcher HA, Donoghue HD, Holton J, Pap I, Spigelman M. Widespread occurrence of Mycobacterium tuberculosis DNA from 18th-19th century Hungarians. Am J Phys Anthropol. 2003;120(2):144–152. | ||
Nagai Y, Iwade Y, Hayakawa E, et al. High resolution melting curve assay for rapid detection of drug-resistant Mycobacterium tuberculosis. J Infect Chemother. 2013;19(6):1116–1125. | ||
Lee ASG, Ong DCT, Wong JCL, Siu GKH, Yam W-C. High-resolution melting analysis for the rapid detection of fluoroquinolone and streptomycin resistance in Mycobacterium tuberculosis. PLoS One. 2012;7(2):e31934. | ||
Lee SH, Choi HB, Yu SY, Chang UJ, Kim CK, Kim HJ. Detection of first-line anti-tuberculosis drug resistance mutations by allele-specific primer extension on a microsphere-based platform. Ann Lab Med. 2015;35(5):487–493. | ||
Jeong YJ, Lee KS, Yim J-J. The diagnosis of pulmonary tuberculosis: a Korean perspective. Precis Future Med. 2017;1(2):77–87. | ||
Chang KC, Yew WW. Management of difficult multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis: Update 2012. Respirology. 2013;18(1):8–21. | ||
Prasad R, Singh A, Balasubramanian V, Gupta N. Extensively drug-resistant tuberculosis in India: Current evidence on diagnosis & management. Indian J Med Res. 2017;145(3):271–293. | ||
Yin X, Zheng L, Liu Q, et al. High-resolution melting curve analysis for rapid detection of rifampin resistance in Mycobacterium tuberculosis: a meta-analysis. J Clin Microbiol. 2013;51(10):3294–3299. | ||
Lee ASG, Ong DCT. Molecular diagnostic methods for the detection of Mycobacterium tuberculosis resistance: the potential of high-resolution melting analysis. Expert Rev Anti Infect Ther. 2012;10(10):1075–1077. | ||
Chang K, Lu W, Wang J, et al. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF assay: A meta-analysis. J Infect. 2012;64(6):580–588. | ||
Lohrasbi V, Talebi M, Bialvaei AZ, et al. Trends in the discovery of new drugs for Mycobacterium tuberculosis therapy with a glance at resistance. Tuberculosis. 2018;109:17–27. | ||
Campbell PJ, Morlock GP, Sikes RD, et al. Molecular Detection of Mutations Associated with First- and Second-Line Drug Resistance Compared with Conventional Drug Susceptibility Testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032–2041. | ||
Pourakbari B, Mamishi S, Mohammadzadeh M, Mahmoudi S. First-Line Anti-Tubercular Drug Resistance of Mycobacterium tuberculosis in IRAN: A Systematic Review. Front Microbiol. 2016;7(9):1139. | ||
Galarza M, Fasabi M, Levano KS, et al. High-resolution melting analysis for molecular detection of multidrug resistance tuberculosis in Peruvian isolates. BMC Infect Dis. 2016;16(1):260. | ||
Haeili M, Fooladi AI, Bostanabad SZ, Sarokhalil DD, Siavoshi F, Feizabadi MM. Rapid screening of rpoB and katG mutations in Mycobacterium tuberculosis isolates by high-resolution melting curve analysis. Indian J Med Microbiol. 2014;32(4):398–403. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.