Back to Journals » International Journal of General Medicine » Volume 16
Molecular Detection of Hemoglobin O-Arab in the Sudanese Population
Authors Elbashir I , Elsayed Yousif TY
Received 3 June 2023
Accepted for publication 28 July 2023
Published 3 August 2023 Volume 2023:16 Pages 3323—3330
DOI https://doi.org/10.2147/IJGM.S421140
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Izzeldin Elbashir,1,2 Tagwa Yousif Elsayed Yousif3
1Department of Medical Laboratory Technology, College of Applied Medical Science, Jazan University, Gizan, Saudi Arabia; 2Faculty of Post Graduate and Scientific Research, Shendi University, Shendi, Sudan; 3Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Jazan University, Gizan, Saudi Arabia
Correspondence: Tagwa Yousif Elsayed Yousif, Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, Jazan University, Gizan, 45142, Saudi Arabia, Email [email protected]
Background and Purpose: Sickle cell disease (SCD) is an inherited hemoglobin disease affecting the red cells and causing hemolytic anemia. It is a very common, endemic disease in Sudan, particularly in the central and western areas of Sudan. Sickle cell anemia (SCA) is when the patient has beta-globin gene variant (Hb S variant). In this study, we screened the co-inheritance of hemoglobin O-Arab mutation among Sudanese sickle cell disease patients.
Study Population and Methods: This cross-sectional study was conducted in the Sudan-Khartoum state from November 2016 to December 2021. Blood samples were collected from a random sample of the known sickle cell disease patients of Kordofan-central Sudan origin. Study-appropriate blood samples were subjected to complete blood count (CBC), hemoglobin capillary electrophoresis (CE) and molecular laboratory investigations. Initial laboratory investigations were done in Sudan, where the DNA sequencing technique was carried out at the Egyptian National Research Center (NRC)-Cairo-Egypt.
Results: The final study’s main results revealed the presence of HB O-Arab genetic mutations among Sudanese Sickle cell disease patients, which estimated to be (5%) co-inherited mutations among our study population (Hb-O Arab; (HBB):c.364G>A (p.Glu122Lys)).
Conclusion: The frequency of Hb-O Arab gene mutations was determined among Sudanese sickle cell disease patients, and the results have shown a (5%) frequency of Hb-O Arab mutation. The study result is the first molecular confirmation of co-inherited Hb-O Arab/sickle cell disease clinical condition in Sudan. The results raise the importance of extended studies of other sickle variant conditions.
Keywords: sickle cell anemia, molecular genetic mutation, Hb O-Arab, Hb-S disease, sickle variants
Introduction
Sickle Cell Disease (SCD)
Sickle cell disease is an inherited chronic hemolytic anemia whose clinical manifestations arise from the tendency of the hemoglobin (HbS or sickle hemoglobin) to polymerize and deform red blood cells into the characteristic sickle shape.1
Hb S is insoluble and crystallizes in environments with minimal oxygen pressure. Deoxygenated sickle hemoglobin can polymerize into long fibers with seven entangled, cross-linked double strands per fiber. Red cell sickling can lead to the blockage of various microcirculation or large vascular segments, resulting in organ infarctions. The substitution of Valine for glutamic acid at position 6 of the protein chain results in the condition known as sickle cell anemia. It has been identified in upwards of one in every four people in West Africa, demonstrating an extremely high pervasiveness rate. Since the carrier state offers some insurance against fatal malaria, this rate has stayed consistent. The DNA that codes for the sixth amino acid in the B-globin chain has undergone a single base alteration (adenine is replaced by thymine), as revealed by the molecular pathology of sickle cell anemia. Sickle cell anemia is the result of this change.2
Sickle Cell Disease Epidemiology
Sickle hemoglobin, also called hemoglobin S, is the most common form of hemoglobinopathy encountered globally. People of African descent and those residing in equatorial Africa are the most likely carriers of the hemoglobin S variant. In some regions of Africa, between 10 and 40% of the population is believed to be heterozygous for the hemoglobin S gene. Approximately 8% of African Americans have sickle cell trait, whereas 1 in 400 to 600 African Americans with the hemoglobin S gene homozygosity have sickle cell anemia. Different concentrations of Hemoglobin S have been identified in Turkey, Saudi Arabia, India, and regions bordering the Mediterranean Sea (Sicily, southern Italy, and northern Greece). Given the prevalence of falciparum malaria in these regions, it is possible that hemoglobin S evolved as a protection against the disease. Genetic research reveals that the sickle mutation appears to have emerged at least four times independently.3
Sickle Cell Diseases Other Than Sickle Cell Anemia (Sickle Variants)
Hemoglobin S heterozygotes, as well as those with another hemoglobinopathy, such as hemoglobin C, hemoglobin D-Los Angeles (also known as D Punjab), hemoglobin O-Arab, or hemoglobin S and -thalassemia, frequently exhibit symptoms and are susceptible to many of the same complications as homozygous sickle cell anemia patients.
People with sickle/hemoglobin C (hemoglobin SC) typically experience a milder form of the disease than those with sickle hemoglobin S homozygosity. Despite this, individuals with sickle/hemoglobin SC are more likely to develop retinopathy, infarcts of the long bones, and pregnancy complications. Instead of the severe splenic infarcts typical of individuals with homozygous sickle cell disease, these individuals typically exhibit moderate splenomegaly. The condition generally is not too extreme in individuals with sickle- + thalassemia, where some normal beta globin chains produced. People with sickle-o thalassemia, in which the thalassemia gene does not make any -globin, have a painful ailment similar to homozygous sickle cell anemia and have a high risk of developing the disease.3
Hb O Arab and O Arab/Sickle Disease
Due to a point mutation from GAA to AAA, the hemoglobin variant known as hemoglobin (Hb), O-Arab has a different amino acid at position 121 of the beta chain (alpha2beta2121Glu Lys, HBBE121K). This results in the modification of an amino acid. The amino acid mutation alters the molecule’s net charge but retains its stability and oxygen-transporting capability. Consequently, the highly positively charged Hb O-Arab molecule accumulates preferentially just beneath the negatively charged inner surface of the erythrocyte membrane. Aggregation causes erythrocytes to become denser and more spherical, which increases MCHC and slightly decreases MCV. It also promotes the polymerization of coexisting sickle hemoglobin. In addition, sickle hemoglobin may be present due to accumulation.
No clinical symptoms are associated with the mutant hemoglobin in heterozygotes with the hemoglobin O-Arab variant. Similarly, homozygotes of Hb O-Arab exhibit moderate hemolysis and borderline splenomegaly, which is clinically significant. This is especially true when neither a severe illness nor an infection is present. Anemia caused by the Hb O-Arab and beta thalassemia genes may or may not require frequent blood transfusions. Similar to patients with Hb Lepore/HbS illness, those with Hb S/Hb O-Arab double heterozygotes exhibit S/S illness symptoms, such as a sickle crisis. Although Hb O-Arab shows a global distribution pattern, it is most prevalent in the Middle East and the Mediterranean. The hilly terrain’s physically and genetically pure Muslim population is known as the Greek Pomaks. They have an astoundingly high proportion of Hb O-Arab in their blood, reaching 27.4% in some regions.4
Materials and Methods
This study is a prospective, cross-sectional study conducted between Kordofan province-central Sudan and Khartoum state-capital of Sudan, between November 2016 and December 2021. The molecular work has been done at the National Research Center (NRC)-in Cairo-Egypt.
This study has been conducted among Sudanese patients from the Kordofan region, which constitutes Sudan’s central and southern areas. It lies between Darfur on the west and the valley of the White Nile on the east.
Random 100 Sudanese sickle cell disease patients from Kordofan province-central Sudan were included in this study.
Inclusion Criteria
Known Sudanese, sickle cell disease patients, based on CBC and hemoglobin electrophoresis screening investigations and clinical features of sickle cell anemia, were included in the study.
Both sexes were included in the study, and there was no limitation regarding the age of the patients, as all age groups of sickle patients were included.
Non-Inclusion Criteria
Non-Sudanese individuals, normal Sudanese individuals and non-sicker patients with negative laboratory and clinical remarks of sickle cell disease were excluded from the study.
Sample Collection
Sample collection was done in the Sickle Anemia Center laboratory in Kordofan state.
Blood samples were taken from known sickle cell disease patients of Kordofan-central Sudan origin.
After cleaning the collection area with 70% alcohol swabs, 3–5 milliliters of venous blood samples were drawn using a disposable plastic syringe and transferred to K2EDTA anti-coagulant sample containers.
Methods and Techniques
Study samples were subjected to initial hematological investigations, including complete blood count (CBC) using Sysmex KX-21N automated hematological analyzer and hemoglobin capillary electrophoresis (CE), using the Capillarys-Minicap from Sebia (Lisses-France).
DNA extraction, PCR, and DNA sequencing were done in the Egyptian National Research Center (NRC) molecular genetics laboratory-Cairo-Egypt.
According to the manufacturer’s instructions, genomic DNA was extracted from peripheral blood samples using QIAamp® DNA Mini Kit (QIAGEN). DNA purity and concentration were evaluated using a NanoDrop™ spectrophotometer and samples stored at −20 °C for molecular analysis.
DNA Sequencing Technique
Sanger Sequencing protocol of Steps & Method was applied for DNA sequencing.5 The total nucleotide succession of the gene encoding beta-globin was amplified utilizing three overlapping primers planned by Primer3 web administration at primer3.ut.ee6 based on the promoter, exonic, and intronic regions of the beta-globin gene (NM_000518) (Table 1).
 |
Table 1 Primer Sequences, Annealing Temperatures, and Lengths of the Amplified Fragments |
One hundred nanograms of DNA, standard PCR buffer, dNTP concentrations, and 1pmoL of each forward and reverse primer were used for PCR amplification.
The PCR regimen was as follows: denaturation at 94°C for 5 minutes, followed by 35 cycles consisting of primer annealing at 58.5°C for 30 seconds, denaturation at 72°C for 30 seconds, and final extension at 72℃ for 5 minutes.
Direct sequencing was done in both directions on amplified fragments. Decontamination of PCR item was finished using QIAquick PCR Purging Pack (Qiagen), agreeing on maker guidelines. Cleaned tests were exposed to cycle sequencing utilizing Enormous Color Eliminator v3.1 Unit and infused to Crystal 3500 Hereditary Analyzer (Applied Bio frameworks, Germany).
The Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov) was used to blast the reference human globin gene on the NCBI gene bank to find HBB mutations using the collected sequencing results.7
Result Analysis & Determination of DNA Sequence
Samples were cycle sequenced using a Big Dye Terminator v3.1 Kit before being inserted into an ABI 3100 Genetic Analyzer (Applied Biosystems-Germany) to determine the sequence. Fluorescence is used to identify each terminal ddNTP in the capillary gel by moving the apparatus from one band to the next in a logical order. A laser stimulates the fluorescent identifiers in each band, and a computer detects the resulting light. It is possible to immediately correlate the emitted light with the identity of the ddNTP serving as the terminal because each of the four ddNTPs has a distinct fluorescent label. This results in a chromatogram that displays each nucleotide’s fluorescence peak along the template DNA’s length.
Data Analysis
After encoding the questionnaire’s data in a primary Excel file, a medical statistician performed the data analysis. The SPSS application, version 11.5, which stands for Statistical Package for the Social Sciences, was used for data entry and analysis. The data were analyzed using descriptive statistics and one-way ANOVA tests to determine the variability of the results.
Ethical Consideration
The study complies with the Declaration of Helsinki and has been approved by the ethical committee of the Faculty of Post Graduate and Scientific Research at Shendi University-Sudan.
Participants were given clear information about the study and asked to sign a written informed consent form before participation.
Results
This study included one hundred patients (42 males, 58 females), as illustrated in Figure 1.
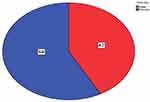 |
Figure 1 Gender distribution of study population. |
The male and female age groups included in the study were ≤5, 5–10 and >10 years old, with male percentages of 44.9%, 35.1% and 50% and female percentages of 55.1%, 64.9% and 50%, respectively (Table 2).
 |
Table 2 Gender and Age Group Distribution of the Study Population |
Molecular Investigation Results
The complete nucleotide sequence of the Beta globulin gene (HBB) was amplified for sequencing using three overlapping primers (Fragment 1, 2 and 3) covering the promoter, exonic and intronic regions (Figure 2).
Results of DNA Sequencing Analysis
Ninety cases (90%) had a homozygous mutation in codon 6 (Hb SS) of the beta-globin gene (HBB): c.20A>T, according to sequencing analysis of the gene’s promoter region and first exon (p. Glu7Val), a heterozygous mutation was carried by 10 cases (or 10%).
Figure 3 depicts the codon’s six normal (wild) and mutant sequences (Figure 3).
Five carriers of the heterozygous mutation in codon 6 (HbS) were also carriers of the Hb O-Arab variant ((HBB):c.364G>A, Glu122 Lys) as shown by further sequencing analysis of intron 1, exon 2, and exon 3 (Table 3 and Table 4 and Figure 4).
 |
Table 3 Hb O-Arab Mutation Screening Results After Sequencing |
 |
Table 4 Hb O-Arab Mutation After HBB Gene Sequencing |
 |
Figure 4 The heterozygous Hb O-Arab variant (HBB):c.364G>A (p.Glu122Lys). Partial chromatogram representing sequencing analysis of intron two and exon 3 of the beta-globin gene. |
Discussion
Sickle cell disease is a common illness in the area of central and west of Sudan, with wide range of serious clinical conditions and genetic variants. As recently stated by Tariq Osman et al (2022) on their geographically similar study, different sickle haplotypes were found among sickle disease Sudanese patients from Kordofan area, the Bantu (BA) sickle haplotype was found in 10.8% of participants with homozygous uncontested haplotypes, followed by Benin (BA) and Sudan (SU), each in 9.8% of participants. The study also revealed that, the Sudanese group from Northern Kordofan lacked the Arab-Indian haplotype and two heterozygous versions of undisputed haplotypes were found in 17.3% of participants: SU/BA in 10.8% and CA/BE in 6.5%.8
In this study and upon beta gene (HBB) entire sequencing, five patients (5%) showed a presence of Hb O-Arab mutation ((HBB):c.364G>A (p.Glu122Lys)).
This molecular detection of Hb O-Arab mutation among Sudanese is considered very unique, as very few previous studies have shown the same finding, which is an obvious confirmation of the special population of S/O-Arab among Sudanese Sickle disease patients.
Hemoglobin O Arab has been previously reported in the literature in northern Sudanese patients in heterozygote states and in combination with sickle hemoglobin.9,10
The historical link for Sudan is that Hb S O-Arab has also been described in North Africa and West Africa among Berbers, as described by El Hazmi et al (1980), Zimmerman et al (1999) and Sangare et al (1992).11–13
A recent Arab study (Ata et al 2023) has represented a systematic and comprehensive review of all the prior published peer-reviewed articles describing genotypes of SCD in the Arabic population. The study captured 44,034 patients diagnosed with SCD in all Arab countries up to November 2020. The review found Hb S O-Arab and HBS Oman in 56 cases and one case, respectively.14
Hemoglobin O-Arab finding in this study is predictable with past studies done in a few Middle Eastern nations and other Center East regions. Old research from Saudi Arabia (El-Hazmi M.A.F) has shown that a new structural abnormality and a new pocket for the sickle-cell gene are the first signs of Hb O-Arab in Saudi Arabia.11
Our momentum concentrates on the consequence of O-Arab change is likewise observed to be viable with a past report done in the US, as 13 African-American kids and grown-ups were related to Hb S/O (Arab) going in age from 2.7 to 62.5 years and presumed that Hb S/O (Arab) illness is an extreme sickling hemoglobinopathy with lab and clinical appearances like those of homozygous sickle cell anemia (S.A Zimmerman).12
We do believe that our study could be an addition to the local and international efforts in hemoglobinopathy study. As described by Ogbonna Collins et al (2019), hemoglobinopathy screening is strongly recommended in policy guidelines of all health institutions. However, the challenge will be on the implementation. This is where the Human Heredity and Health in Africa (H3Africa) Initiative and sickle cell genomic network of Africa should make their impact in the health institutions.15
Conclusion
The entire sequencing of the study population’s beta globulin gene has revealed the detection and the confirmation of the Sudanese S/O-Arab mutations ((HBB):c.364G>A), which is estimated to be 5% in our study.
In conclusion, these new mutation detections account for a good record for the group of hemoglobinopathies and Sickle variants in Sudan and would thoughtfully help in Sickle disease management.
Recommendations
Our study was one of the very few molecular studies of sickle variants in Sudan. We recommend continuing the molecular genetic work among Sickle disease Sudanese patients, covering all sickle variants and Alpha globulin chain genetic screening.
Implementing molecular and clinical research through hospital-based coherent studies or projects would assure a successful outcome, and eventually good disease management.
The importance of the neonatal molecular screening program and genetic counseling should be raised, as it surely will result in early sickle variant disclosure and optimum disease control. Since there are no clear registry data about Sickle disease and the exact sickle population in Sudan, we recommend a national screening research or project and establishing a governing body or a special authority for this purpose.
Sickle cell newborn screening programs could be built upon Point-Of-Care tests such as Hemex Gazelle, which has been reported and indicted by Kelli Qua et al (2021).16
Strengths and Limitations
Our study had several strengths. The research topic and idea considered to be unique according to previous studies in the study area and the large size study population were the main strengths.
This study had two main limitations. Because the clinical condition related to the research topic is rare, and due to lack of official disease population data and previous similar molecular studies on the research topic, the first main limitation was the lack of sufficient data about the study population and the research topic, which made some deficiency in the theoretical and methodological foundations for the study. The second limitation was concerning the sampling time, due to the rareness and the special localization of the study population, sampling duration time was very long, which made the study to be completed over a long period of time.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Victor Hoffbrand A, Catovsky D, Edward GD. Postgraduate haematology.
2. Hoffbrand AV, Moss PAH, Pettit JE. Essential Hematology.
3. William F, Kern MD. PDQ Hematology.
4. Papadopoulos V. The implications of hemoglobin O-Arab mutation. Haematologica. 2003; 2003:1.
5. Merck KG. Sanger sequencing steps & method: Germany; 2021.
6. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi:10.1093/nar/gks596
7. Altschul SF, Gish W, Miller W, Myers E, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi:10.1016/S0022-2836(05)80360-2
8. Khalafallah TO, Ajab Eldoor AA, Babker AM, et al. Hematological and molecular analyses of the HbS allele among the Sudanese population. J Int Med Res. 2022;50(9):3000605221125050. PMID: 36134571; PMCID: PMC9502246. doi:10.1177/03000605221125050
9. Ibrahim SA, Mustafa D. Sickle-cell haemoglobin O disease in a Sudanese family. Br Med J. 1967;3(5567):715–717. PMID: 6038366; PMCID: PMC1843069. doi:10.1136/bmj.3.5567.715
10. Vella F, Beale D, Lehmann H. Haemoglobin O Arab in Sudanese. Nature. 1966;209(5020):308–309. PMID: 5915974. doi:10.1038/209308a0
11. ElHazmi MAF, Lehmann H. Human haemoglobins and haemoglobinopathies in Arabia: Hb O Arab in Saudi Arabia. Acta Haematol. 1980;63:268–273. doi:10.1159/000207414
12. Zimmerman SA, O’Branski EE, Rosse WF, Ware RE. Hemoglobin S/O (Arab): thirteen new cases and review of the literature. Am J Hematol. 1999;60:279–284. doi:10.1002/(SICI)1096-8652(199904)60:4<279::AID-AJH5>3.0.CO;2-2
13. Sangare A, Sanogo I, Meite M, et al. L’hémoglobine o Arab en Côte d’Ivoire et en Afrique de l’Ouest [Hemoglobin O Arab in Ivory Coast and western Africa]. Med Trop. 1992;52(2):163–167. French. PMID: 1328806.
14. Ata F, Rahhal A, Malkawi L, et al. Genotypic and phenotypic composition of sickle cell disease in the Arab population - a systematic review. Pharmgenomics Pers Med. 2023;16:133–144. PMID: 36851992; PMCID: PMC9961577. doi:10.2147/PGPM.S391394
15. Collins Nwabuko O, Eke R, Jean Claude Kazadi M. Beyond the legislation for sickle cell disease prevention - getting the right outcomes. Am J Intern Med. 2019;7(3):56–65. doi:10.11648/j.ajim.20190703.12
16. Qua K, Swiatkowski SM, Gurkan UA, Pelfrey CM. A retrospective case study of successful translational research: gazelle Hb variant point-of-care diagnostic device for sickle cell disease. J Clin Transl Sci. 2021;5(1):e207. doi:10.1017/cts.2021.871
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


