Back to Journals » Infection and Drug Resistance » Volume 12
Molecular characterization of the pilS2 gene and its association with the frequency of Pseudomonas aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island
Authors Bahramian A , Khoshnood S, Shariati A , Doustdar F, Salimi Chirani A, Heidary M
Received 1 October 2018
Accepted for publication 12 December 2018
Published 11 January 2019 Volume 2019:12 Pages 221—227
DOI https://doi.org/10.2147/IDR.S188527
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Aghil Bahramian,1,2 Saeed Khoshnood,3,4 Aref Shariati,5 Farahnoosh Doustdar,2 Alireza Salimi Chirani2 Mohsen Heidary5,6
1Infectious Diseases and Tropical Medicine Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 3Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 4Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 5Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran; 6Student Research Committee, School of Allied Medical Sciences, Iran University of Medical Sciences, Tehran, Iran
Introduction: Pseudomonas aeruginosa is the most common opportunistic pathogen associated with a broad range of infections, including cystic fibrosis, ocular, otitis media, and burn infections. The aim of this study was to show the frequency of the pilS2 gene, and its association with P. aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island among P. aeruginosa strains.
Methods: The samples were collected from patients with cystic fibrosis, ocular, otitis media, and burn infections between January 2016 and November 2017. DNA was extracted using the DNA extraction kit and was used for PCR assay. PCR with 4 primer-pairs including 976 F/PAPI-1R, 4542 F/intF, SojR/4541 F, and intF/sojR was performed to identify PAPI-1. pKLC102 was detected using three other primer-pairs including cp10F/ cp10R, cp44F/cp44R, and cp97F/cp97R.
Results: A total of 112 P. aeruginosa isolates were collected from patients with cystic fibrosis (36), burn (20), otitis media (26), and ocular (30) infections. The results of PCR showed that pilS2 gene was identified in 96 (85%) strains. PAPI-1–attB integration was detected among 38 (33.9%) isolates and the circular form of PAPI-1 detected among 17 (14%) isolates. In addition, 79 (70.5%) strains were found to be positive for pKLC102.
Conclusion: We found that the majority of the isolates may be susceptible to transfer this significant island and the related element pKLC102 into recipient isolates lacking the island owing to high association of the PilS2 pilus with the islands in the studied strains. It is anticipated that strains isolated from burn and eye with the highest rate of PilS2, PAPI-1, and pKLC102 association have a high level of antibiotic resistance.
Keywords: Pseudomonas aeruginosa, pilS2, cystic fibrosis, pKLC102, PAPI-1
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is the most common bacterial pathogen associated with persistent bronchial infections among cystic fibrosis patients.1 This pathogen is capable of causing several infections including nosocomial pneumonia, nosocomial urinary tract infections, acute diffuse otitis media, conjunctivitis, and keratitis.2–4 The pathogenesis of P. aeruginosa is due to the production of extracellular virulence factors (such as pili, hemolysins, pyocyanin, proteases, alginate, lipopolysaccharide, and the Type III secretion system) and effector proteins (such as ExoU, ExoT, ExoY, and ExoS).5,6
P. aeruginosa isolates have shown the ability of adherence to different receptors and production of biofilm by Type IV pili.7 Type IV pili has two major subfamilies including Type IVa (T4aP) and IVb (T4bP). T4bP are a more heterogeneous group, whereas T4aP are a somewhat homogeneous and Tad pili (a monophyletic class of T4bP) were seen in a broad variety of environmental species.8
Although T4aP- and Tad-encoding genes are found in all the P. aeruginosa isolates, the gene encoding major subunit of the Type IVb pili (pilS2 gene) is seen in the strains that have related elements such as P. aeruginosa plasmid pKLC102 or PAPI-1 pathogenicity island.8,9 Carter et al showed that PAPI-1 can be transferred to other P. aeruginosa strains following excision from the chromosome of the donor. Also, they demonstrated that PAPI-1 is transferred into recipient P. aeruginosa by a conjugative mechanism, via a Type IV pilus, encoded in PAPI-1 by a ten-gene cluster (8).
There are two tRNALys genes located in the P. aeruginosa genome, known as PA0976.1 and PA4541.1. pKLC102 is a 103,532 bp integrative and conjugative element that integrates into one of these positions.10 However, PAPI-1 can integrate into the attB sites of the both PA0976.1 and PA4541.1 positions.11 In addition to this integrated form of PAPI-1, the island can also exist in some populations of bacteria as an extrachromosomal circular form.12
Interestingly, the 10 PAPI-1-encoded pilus proteins are well conserved in several P. aeruginosa strains that carry this island, including PA2192, PA7, C3710, PACS2, and PSE9 (PAGI-5), and in P. aeruginosa clone C strain that carry a pKLC102-like element.
The PAPI-1 pilS2 gene encodes for a major pilin subunit, which is a 176-amino-acid protein containing a conserved PilS superfamily domain. Horizontal gene transfer is known as a main evolutionary mechanism and contributes to the virulence properties of many bacterial pathogens (V).
There is no specific study on the prevalence of the pilS2 gene and its association with P. aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island among P. aeruginosa in Iran. This study could be the basis for further research in the future.
So, the aim of this study was to evaluate the prevalence of the pilS2 gene, and its association with P. aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island among P. aeruginosa strains isolated from patients with cystic fibrosis, ocular, otitis media, and burn infections.
Methods
Study design, data, and specimens collection
A prospective cross-sectional study was conducted between January 2016 and November 2017. The research was approved by the ethical committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran with ethics number: IR.SBMU.RETECH.REC.1395.460. As a part of the Shahid Beheshti University of Medical Sciences policy, written informed consent was obtained from all patients. The study was conducted in accordance with the Declaration of Helsinki. The clinical samples were collected from inpatients with otitis media, burn wound infections, keratitis, and cystic fibrosis who were hospitalized in Shahid Motahari Hospital (level I burn care center). The sputum samples of the patients with cystic fibrosis were examined macroscopically for the presence of salivary contamination. Following this, briefly, sputum was incubated with an equal volume of sputolysin for 15 minutes at room temperature and plated onto chocolate agar. The deep oropharyngeal swab samples were collected and cultured on blood agar and MacConkey agar media (Merck, Germany).13
For bacterial isolation from the burn patients, surface culture swabs were collected from them and were inoculated onto blood agar, MacConkey agar, and tryptic soy broth media (Merck, Germany).14 The ocular specimens from the patients with conjunctivitis and keratitis were collected by an ophthalmic surgeon and ophthalmologist. These specimens were transferred into 2 mL of brain-heart infusion broth (Oxoid, Hampshire, UK).15
The outer ear of the patients with otitis media was cleaned with normal saline and the pus discharges were collected with sterile cotton swab sticks. Pus swabs were added to stuart transport medium (Merck, Germany) and cultured on blood agar, chocolate agar, and MacConkey agar media (Merck, Germany).16
The isolated colonies from the positive cultures were investigated for biochemical characteristics of P. aeruginosa. Briefly, the biochemical tests include gram staining, pigment production, colony morphology, growth at 42°C on Mueller–Hinton agar, the ability of oxidase and catalase production, and oxidative-fermentative test.1
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the Kirby–Bauer disk diffusion method according to Clinical & Laboratory Standards Institute17 recommendations. The susceptibilities of all isolates to different antibiotics were determined using cefepime (30 µg), amikacin (30 µg), ceftazidime (30 µg), tobramycin (10 µg), ciprofloxacin (5 µg), meropenem (10 µg), cefoperazone (75 µg), piperacillin (100 µg), imipenem (10 µg), gentamicin (10 µg), ceftazidime (30 µg), piperacillin-tazobactam (110/10 µg), colistin (10 µg), cefoperazone-sulbactam (100/10 µg), and cotrimoxazole (10 µg).
DNA extraction and PCR
The clinical samples were cultured on chocolate agar medium and the isolated single colonies of P. aeruginosa were incubated in 5 mL Luria–Bertani medium at 37°C overnight. Then, DNA templates were prepared using the DNA extraction kit (Genet Bio Company, Korea, Cat. No, K-3000) as previously described.18
PCR assay with four primer-pairs including 976F/PAPI-1R, 4542F/intF, SojR/4541F, and intF/sojR was performed to identify the P. aeruginosa PAPI-1 pathogenicity island.
Likewise, P. aeruginosa plasmid pKLC102 was detected using three other primer-pairs including cp10F/ cp10R, cp44F/cp44R, and cp97F/cp97R by PCR method. Primer sequence used in the genetic characterization of P. aeruginosa clinical strains and for detecting the pilS2 gene is shown in Table 1. Amplified products were analyzed by electrophoresis on 1% agarose gel containing safe stain, and finally in the case of DNA replication and seeing the sharp band, it is considered as positive.
  | Table 1 Primer sequence used in the genetic characterization of Pseudomonas aeruginosa clinical strains |
Data analysis
The data were analyzed using the SPSS statistical package version 22 (IBM SPSS Statistics, USA) and Microsoft excel 2016 (Microsoft Corporation, USA) statistical software. The results are presented as descriptive statistics in terms of relative frequency.
Results
A total of 112 P. aeruginosa isolates were collected from different sources, of which 36 were isolated from patients with cystic fibrosis, 20 from burn wound infections, 26 from otitis media, and 30 from ocular infections.
Antibiotic susceptibility tests using the Kirby–Bauer method revealed that the highest resistance percentage was related to cefepime, gentamicin, and ciprofloxacin 99 (88%), followed by meropenem and imipenem 98 (87%). The results showed that colistin and cefoperazone-sulbactam were the best antibiotics against P. aeruginosa isolates. Antibiotic resistance patterns are detailed in Table 2.
  | Table 2 Antibiotic resistance patterns in Pseudomonas aeruginosa isolates |
Frequency of the pilS2 gene
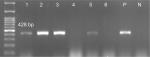
The results showed that the pilS2 gene was identified in 85% (n=96) of the P. aeruginosa strains. This major subunit of Type IVb pili was detected among 24% (n=23) of the P. aeruginosa strains isolated from cystic fibrosis patients, 21% (n=20) isolated from burn patients, 31% (n=30) isolated from patients with ocular infections, and 24% (n=23) isolated from patients with otitis media. Agarose gel electrophoresis of PCR products after amplification of the pilS2 gene is shown in Figure 1.
Frequency of the P. aeruginosa PAPI-1 pathogenicity island
Overall, the integration of the PAPI-1 to the attB site of positions PA0976.1 and PA4541.1 was identified among 33.9% (n=38) of P. aeruginosa isolates. Approximately 18.5% (n=7) of the strains were isolated from cystic fibrosis patients, 39.5% (n=15) isolated from burn patients, 21% (n=8) isolated from patients with ocular infections, and 21% (n=8) isolated from patients with otitis media.
In addition, the PCR assay using primer-pair intF/sojR has detected the circular form of PAPI-1 among 14% (n=17) of P. aeruginosa strains isolated from hospitalized patients (including six cystic fibrosis patients, six burn patients, and five patients with ocular infections).
Frequency of the P. aeruginosa plasmid pKLC102
Overall, 70.5% (n=79) of the P. aeruginosa strains were found to be positive for pKLC102. The highest frequency of this element was detected among patients with ocular (34%) and burn (25%) infections, followed by cystic fibrosis (22%) and otitis media (19%). The results of PCR method for identification of the pKLC102 demonstrated that CP10, CP44, and CP97 genes were present in 49%, 51%, and 17% of the P. aeruginosa strains, respectively (Table 2). Agarose gel electrophoresis of PCR products after amplification of the CP44 and CP10 genes is shown in Figures 2 and 3.
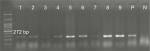  | Figure 2 Detection of CP44 gene (272 bp) in Pseudomonas aeruginosa isolates. Note: Molecular pattern: 100 bp. Abbreviations: N, negative control; P, positive control. |
  | Figure 3 Detection of CP10 gene (255 bp) in Pseudomonas aeruginosa isolates. Note: Molecular pattern: 100 bp. Abbreviations: N, negative control; P, positive control. |
The association of the pilS2 gene with pKLC102
The pilS2 gene was simultaneously associated with pKLC102 in 75 (70%) isolates, of which 27 (36%), 19 (25.4%), 15 (20%), and 14 (18.6%) were detected from ocular, burn, otitis media, and cystic fibrosis infections, respectively (Table 3).
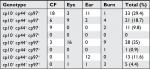  | Table 3 Frequency of the specific genes CP10, CP44, and CP97 of pKLC102 Abbreviation: CF, Cystic fibrosis. |
The association of the pilS2 gene with PAPI-1 and pKLC102
In this study, 36 (32.1%) P. aeruginosa strains encoded all pKLC102, chromosomal PAPI1, plasmid PAPI1, and pilS2 genes. Moreover, the pKLC102+, chromosomal PAPI1–, plasmid PAPI1+, pilS2+ genotype was the most frequent among the isolates (Table 4).
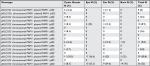  | Table 4 The association of pilS2 with pKLC102, chromosomal PAPI-1, and plasmid PAPI-1 |
The association of the pKLC102 with PAPI-1
The results showed that all the P. aeruginosa strains that are pKLC102 positive encoded also the PAPI-1 element. Although among 37 (33%) strains, pKLC102 was associated with both the chromosomal and plasmid PAPI-1, in 41 (36.6%) strains, pKLC102 was present along with only the plasmid form of the PAPI-1 (Table 5).
  | Table 5 The association of pKLC102 with chromosomal PAPI-1 and plasmid PAPI-1 |
Discussion
P. aeruginosa is an opportunistic human pathogen capable of causing a broad range of human life-threatening acute and chronic infections.19 The bacterium is capable of inhabiting a wide range of environmental niches, in part, owing to have a diverse genomic repertoire that encodes multiple virulence factors such as Type IVb pilus, which is encoded by the island PAPI-1.6 Also, this pathogen can adhere to different receptors and produce biofilm by its extracellular virulence factor, Type IV pili.20
The current study showed the frequency of the gene encoding major subunit of the Type IVb pili, the pilS2 gene, and its association with P. aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island among P. aeruginosa strains isolated from patients with cystic fibrosis, keratitis, otitis media, and burn infection. The results demonstrated that 85% of the P. aeruginosa strains encoded the pilS2 gene. Indeed, 21% (n=20) and 31% (n=30) of the P. aeruginosa strains isolated from patients with burn and ocular infections were pilS2-positive. Furthermore, patients with otitis media (88.4) and cystic fibrosis (63.8) also had a high prevalence of the pilS2 gene.
These findings are comparable with the study performed by Dabiri et al,14 in Iran. They evaluate the Type IV pilin subgroups in P. aeruginosa strains and showed that pilS2 gene was found among 67.5% of cystic fibrosis and 73.3% of burn patients. In this manner, regarding the importance of Type IV pili in virulence and pathogenesis of P. aeruginosa, the results highlight the need for a main program for identification of this pilin and urgently interventions to prevent the development of P. aeruginosa strains isolated from hospitalized patients.
In a study by Sadeghifard et al21 in 2012, the existence of PAPI-1 in clinical P. aeruginosa isolates was evaluated; in that study, 35% of isolates were positive for this genomic island, so that most of the PAPI-1-positive isolates showed high levels of antibiotic resistance. In another study done by Morales-Espinosa et al in Mexico, the genetic and phenotypic features of 100 P. aeruginosa isolates were studied. They have detected PAPI-1 among 81% of the isolates. This island was reported to be integrated into the chromosome at locus PA0976.1 and PA4541.1 among 65% and 16% of the strains, respectively. However, the circular form of PAPI-1 was not found in any of the strains.22
In addition, in Morales-Espinosa et al’s work, 87% of isolates carried pKLC102, so that CP10, CP44, and CP97 were positive in 87, 88, and 70 strains, respectively, and more than half of the isolates carried all three genes. Also, Klockgether et al found 50 isolates of P. aeruginosa with pKLC102-like islands in their study.
Similarly, we reported that the prevalence of integrated form of PAPI-1 was 33.9%. This rate was the highest among patients with burn infection (39.5%), followed by ocular (21%), otitis media (21%), and cystic fibrosis (18.5%) infections. Interestingly, all isolates positive for pKLC102 in the current study were also shown to carry PAPI-1 (in 70.5% of the isolates) and this association was more frequently detected among eye and burn isolates (Table 4).
Furthermore, as our results revealed, the association of pilS2 gene with PAPI-1 and pKLC102 was found predominantly in P. aeruginosa isolates from burn (pKLC102+, chromosomal PAPI1+, plasmid PAPI1+, and pilS2+ genotype) and eye infection (pKLC102+, chromosomal PAPI1, plasmid PAPI1+, and pilS2+ genotype) (Table 4); taken together, in 70.8% of the isolates, the association was established and in 9.8% of the isolates pilS2 gene was only associated with PAPI-1 (chromosomal PAPI-1).
Conclusion
In this study, we found that the majority of the isolates may be susceptible to transfer this significant island and the related element pKLC102 into recipient isolates lacking the island owing to high association of the PilS2 pilus with the islands in the studied strains, which may affect the pathogenesis of these isolates.
Moreover, based on this data, it is anticipated that strains isolated from burn and eye with the highest rate of PilS2, PAPI-1, and pKLC102 association have a high level of antibiotic resistance and a higher potential of biofilm formation. Regarding this, more investigations are needed to evaluate the frequency of PilS2, PAPI-1, and pKLC102 and its association with antibiotic-resistance properties and biofilm synthesis in P. aeruginosa isolates from Iran.
Acknowledgments
We would like to thank the Department of Microbiology, Faculty of Medicine, Shahid Beheshti University of Medical Sciences for their cooperation. Our appreciation goes to the Vice Chancellor for Research Affairs, Shahid Beheshti University of Medical Sciences, Tehran, Iran, and the Tropical and Infectious Diseases Research Center of the university for their financial and executive support.
Disclosure
The authors report no conflicts of interest in this work.
References
Heidary M, Hashemi A, Goudarzi H, et al. The antibacterial activity of Iranian plants extracts against metallo beta-lactamase producing Pseudomonas aeruginosa strains. J Paramed Sci. 2016;7(1):13–19. | ||
Wang MC, Liu CY, Shiao AS, Wang T. Ear problems in swimmers. J Chin Med Assoc. 2005;68(8):347–352. | ||
Buchele MLC, Wopereis DB, Casara F, et al. Contact lens-related polymicrobial keratitis: Acanthamoeba spp. genotype T4 and Candida albicans. Parasitol Res. 2018;117(11):3431–3436. | ||
Tarashi S, Heidary M, Dabiri H, Nasiri MJ. Prevalence of drug-resistant Pseudomonas aeruginosa in Iranian burned patients: a meta-analysis. Arch Trauma Res. 2017;6(3):1–17. | ||
Chirani AS, Majidzadeh R, Pouriran R, et al. The effect of in silico targeting Pseudomonas aeruginosa patatin-like protein D, for immunogenic administration. Comput Biol Chem. 2018;74:12–19. | ||
Harrison EM, Carter ME, Luck S, et al. Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect Immun. 2010;78(4):1437–1446. | ||
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015(1):1–17. | ||
Carter MQ, Chen J, Lory S. The Pseudomonas aeruginosa pathogenicity island PAPI-1 is transferred via a novel type IV pilus. J Bacteriol. 2010;192(13):3249–3258. | ||
Würdemann D, Tümmler B. In silico comparison of pKLC102-like genomic islands of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2007;275(2):244–249. | ||
Klockgether J, Reva O, Larbig K, Tümmler B. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186(2):518–534. | ||
Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. | ||
Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103(52):19830–19835. | ||
Huus KE, Joseph J, Zhang L, et al. Clinical isolates of Pseudomonas aeruginosa from chronically infected cystic fibrosis patients fail to activate the inflammasome during both stable infection and pulmonary exacerbation. J Immunol. 2016;196(7):3097–3108. | ||
Dabiri H, Nikokar I, Dousdar F, Goudarzi H, Fallah F, Esmaili A. Evaluation of type IV pilin sub groups in Pseudomonas aeruginosa isolated from environmental samples, cystic fibrosis and burn patients. Iran J Med Microbiol. 2014;8(3):1–7. | ||
Teweldemedhin M, Saravanan M, Gebreyesus A, Gebreegziabiher D. Ocular bacterial infections at Quiha Ophthalmic Hospital, Northern Ethiopia: an evaluation according to the risk factors and the antimicrobial susceptibility of bacterial isolates. BMC Infect Dis. 2017;17(1):207. | ||
Seid A, Deribe F, Ali K, Kibru G. Bacterial otitis media in all age group of patients seen at Dessie referral hospital, North East Ethiopia. Egypt J Ear Nose Throat Allied Sci. 2013;14(2):73–78. | ||
Hosseini SM, Arabestani MR, Mahmoodi H, Farhangara E. Prevalence of G, H, I, J. Enterotoxin genes and antibacterial susceptibility pattern in staphylococcus aureus strains isolated from different foods. Middle East J Fam Med. 2015;13(4):1–10. | ||
Khoshnood S, Eslami G, Hashemi A, et al. Distribution of aminoglycoside resistance genes among Acinetobacter baumannii strains isolated from burn patients in Tehran, Iran. Arch Pediatr Infect Dis. 2017;5(3):e577263. | ||
He J, Baldini RL, Déziel E, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA. 2004;101(8):2530–2535. | ||
Rasamiravaka T, Vandeputte OM, Pottier L, et al. Pseudomonas aeruginosa biofilm formation and persistence, along with the production of quorum sensing-dependent virulence factors, are disrupted by a triterpenoid coumarate ester isolated from Dalbergia trichocarpa, a tropical legume. PLoS One. 2015;10(7):e0132791. | ||
Sadeghifard N, Rasaei SZ, Ghafourian S, et al. Prevalence of genomic island PAPI-1 in clinical isolates of Pseudomonas aeruginosa in Iran. Southeast Asian J Trop Med Public Health. 2012;43(2):431–435. | ||
Morales-Espinosa R, Soberón-Chávez G, Delgado-Sapién G, et al. Genetic and phenotypic characterization of a Pseudomonas aeruginosa population with high frequency of genomic islands. PLoS One. 2012;7(5):e37459. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.