Back to Journals » Infection and Drug Resistance » Volume 16
Molecular Characteristics, Antimicrobial Susceptibility, Biofilm-Forming Ability of Clinically Invasive Staphylococcus aureus Isolates
Authors Wang W, Zhong Q, Cheng K, Tan L, Huang X
Received 25 September 2023
Accepted for publication 7 December 2023
Published 18 December 2023 Volume 2023:16 Pages 7671—7681
DOI https://doi.org/10.2147/IDR.S441989
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Weiguo Wang, Qiuxaing Zhong, Ke Cheng, Lili Tan, Xincheng Huang
Department of Clinical Laboratory, The First Hospital of Nanchang, Nanchang, Jiangxi, People’s Republic of China
Correspondence: Xincheng Huang, Email [email protected]
Purpose: This study aimed to investigate the molecular characteristics, antimicrobial resistance, and biofilm-forming ability of Staphylococcus aureus isolates from invasive infections.
Methods: A total of 92 non-repetitive S. aureus isolates from invasive infections were analyzed by Multi-locus Sequence Typing (MLST), spa typing, and chromosomal cassette mec (SCCmec) typing. Antibiotic susceptibility testing was performed using the disk diffusion and agar dilution methods. Biofilm-forming ability was assessed using crystal violet assay. The presence and expression of biofilm-associated genes were examined using PCR and RT-qPCR.
Results: Among the 55 Methicillin-resistant S. aureus (MRSA) and 41 Methicillin-sensitive S. aureus (MSSA) isolates, ST59 (43.6%) predominated in MRSA, while ST7 (39.0%) was most common in MSSA. As expected, MRSA exhibited higher antibiotic resistance rates compared to MSSA isolates. Biofilm formation assays revealed that the majority of isolates (88.5%) produced biofilms, with 26.0% classified as strong producers (OD570 ≥ 1.0) and 62.5% as weak producers (0.2 ≤ OD570< 1.0). MSSA exhibited a higher biofilm-forming ability than MRSA (P < 0.01), with variations across clones. Notably, ST7 isolates displayed greater biofilm-forming ability than other sequence types (ST59, ST5, and ST239). RT-qPCR results revealed that ST7 isolates exhibited higher expression levels of icaA compared to other sequence types.
Conclusion: This study revealed significant molecular heterogeneity among invasive S. aureus isolates, with ST59 and ST7 as dominant clones. The strong biofilm-forming capacity of ST7 merits concern given its rising prevalence regionally. Continuous surveillance of emerging successful lineages is critical to help guide infection control strategies against invasive S. aureus infections.
Keywords: Staphylococcus aureus, invasive infection, biofilm, ST7
Corrigendum for this paper has been published.
Introduction
Staphylococcus aureus is a major bacterial pathogen capable of causing infections ranging from mild skin infections to life-threatening invasive diseases.1,2 The emergence of methicillin-resistant S. aureus (MRSA), especially in healthcare settings, has raised global concerns due to its multidrug resistance and its association with invasive infections. The treatment of invasive MRSA infections is further complicated by the continued acquisition of antibiotic resistance determinants.
Among the critical virulence factors of S. aureus, biofilm formation takes center stage as a pivotal contributor to its pathogenicity. Biofilms are structured assemblies of surface-associated microbial cells encased within an extracellular polymeric substance matrix.3 These intricate microbial communities frequently establish themselves on various medical devices, such as intravascular catheters, prosthetic heart valves, and orthopedic implants.4 Bacteria residing within biofilms exhibit enhanced resistance to antimicrobial agents through a combination of mechanisms.5 The penetration of antimicrobials into the biofilm is hindered, growth-dependent agents become less effective due to reduced metabolic activity, and the proximity of cells facilitates the exchange of resistance genes.5 Additionally, the formation of persister cells, a subpopulation capable of surviving antimicrobial treatments, further compounds the challenge.6 Consequently, biofilm-related infections on prosthetic devices can serve as persistent reservoirs and trigger metastatic complications, including endocarditis, deep-seated abscesses, septic arthritis, and osteomyelitis.5 Management often necessitates the removal or replacement of the infected prosthetic material.7
Molecular typing techniques including multilocus sequence typing (MLST), spa typing, and SCCmec typing, provide critical insights into the genetic diversity and clonal distribution of S. aureus populations, including MRSA.8 Characterizing the predominant lineages, virulence factors, antibiotic resistance profiles, and biofilm-forming capabilities of S. aureus isolates from invasive infections informs effective infection control and therapeutic strategies.
This study aims to comprehensively characterize the genetic profiles, antimicrobial resistance phenotypes, and biofilm-forming ability of S. aureus isolates obtained from invasive infections.
Methods
Bacterial Isolates
Invasive S. aureus infections represent a critical infection, characterized by the isolation of S. aureus from typically sterile regions within the human body (including blood, bones, joint/synovial fluid, pleural fluid, and peritoneal fluid). From September 2020 to September 2022, 96 invasive S. aureus isolates (single isolate per patient) were collected from the First Hospital of Nanchang, China. To identify these isolates, we cultured the samples on blood agar plates and incubated them at 37°C in a 5% CO2 environment for 24 hours. Subsequently, the VITEK-2 Compact was employed for bacterial identification. S. aureus ATCC25923 was used as a control strain.
Antimicrobial Susceptibility Testing
S. aureus susceptibility to penicillin, gentamicin, clindamycin, erythromycin, tetracycline, ciprofloxacin, rifampicin, and linezolid was assessed using the disc diffusion test method recommended by the Clinical and Laboratory Standards Institute (CLSI). Oxacillin and vancomycin minimum inhibitory concentrations for S. aureus isolates were determined through the broth microdilution methodology. Strains of S. aureus ATCC 25923 and Escherichia coli ATCC 25922 were employed as quality controls in the study.
DNA Extraction, MLST Typing, MRSA Identification, spa Typing, and SCCmec Typing
DNA extraction was carried out using the Rapid Bacterial Genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s protocol. MLST was conducted by amplifying internal fragments of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) of S. aureus, as previously described.9 The obtained PCR product sequences were then compared to the existing alleles in the MLST database (http://saureus.mlst.net/), which assigned a specific allelic number, known as a sequence type (ST), to each isolate. Amplification of the spa gene was also performed using previously described primers.10,11 The PCR products were sequenced by Tsingke Biological Technology Company (TsingKe, Beijing, China), and the spa types were determined using the Ridom SpaServer (http://www.spaserver.ridom.de). The identification of MRSA was initially carried out through cefoxitin screening, with confirmed cases being verified by the presence of the mecA gene using PCR (F: 5’-3’ GTGAAGATATACCAAGTGATT; R: 5’-3’ ATGCGCTATAGATTGAAAGGAT).12 SCCmec typing of MRSA isolates was conducted using a battery of multiplex PCRs, as previously described.13 MRSA isolates that displayed unanticipated fragments or lacked fragments in the multiplex PCR were categorized as non-typeable (NT).
Biofilm Semi-Quantitative Assay
The semi-quantitative biofilm assay was conducted using 96-well tissue culture plates, following the method described by Xiao et al.14 In brief, S. aureus cells cultured overnight in Tryptic Soy Broth (TSB) at 37°C were diluted 200-fold. Subsequently, 200 µL of the diluted culture was transferred to individual wells of a 96-well polystyrene microtiter plate. After 24 hours of incubation at 37°C, the wells were washed three times with phosphate-buffered saline to remove planktonic bacteria. Each well was then fixed with 100 µL of 100% methanol to stabilize the biofilm for 10 minutes, followed by staining with 100 µL of crystal violet for an additional 10 minutes. The excess stain was gently washed off under slowly running water, and the plates were air-dried at room temperature. Subsequently, crystal violet was dissolved in each well with 200 µL of 30% acetic acid. The absorbance of the stained biofilm was measured at 570 nm using a Synergy2 (Bio-Tek) microplate reader. Standard S. aureus ATCC 25923 (a strong biofilm producer) and ATCC 12228 (a non-biofilm producer) were included in the study as reference strains. Biofilm formation was interpreted as strongly positive (OD570 ≥ 1.0), weakly positive (0.2 ≤ OD570 < 1.0), or negative (OD570 < 0.2). All experiments were conducted in triplicate.
Detection of Biofilm Related Genes
The presence of various biofilm-related genes was assessed through a conventional PCR assay targeting the following ten genes: polysaccharide intercellular adhesions (icaA and icaD), fibronectin-binding proteins A and B (fnbA, fnbB), clumping factors A and B (clfA, clfB), ebps, sdrC, sdrD, and sdrE. PCR primers employed for these assays are detailed in Table 1. Amplification was conducted using an ABI ProFlex thermal cycler (Applied Biosystems, USA). Each PCR reaction included a negative control (distilled water). The PCR fragments were visualized by 1.5% agarose gel electrophoresis and ethidium bromide staining.
 |
Table 1 PCR Primers are Used for PCR Assays |
Real-Time Quantitative PCR (RT-qPCR)
S. aureus isolates were cultured in TSB at 37°C for 9 hours. Total RNA was extracted using the Sangon Biotech RNA purification kit. The quality and integrity of total RNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, MA, USA) and agarose gel electrophoresis. Reverse transcription was carried out using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara) following the manufacturer’s protocol. RT-qPCR amplification reactions were prepared in a final volume of 20 μL, containing 400 ng of cDNA, 10 μL of TB GreenTM premix (Takara), 0.5 μL each of forward and reverse primers (10 nM each), and nuclease-free water to reach the final volume of 20 μL. The primer sequences used for icaA are provided in Table 1. The relative RNA expression changes of target genes were calculated using the 2−ΔΔCt formula.15 RT-qPCR was performed on a QuantStudioTM 5 Real-Time PCR System, and each reaction was run in triplicate.
Statistical Analysis
Statistical analysis and data visualization were performed using GraphPad Prism 8.0 software. For the analysis of categorical data, we utilized χ2 and Fisher’s exact tests as deemed appropriate. Unpaired two-tailed Student’s t-tests were conducted to evaluate the statistical significance of biofilm formation in S. aureus isolates. A p-value of less than 0.05 was considered statistically significant.
Results
Characteristics and Diversity of S. aureus Isolates from Invasive Infections
In this study, a total of 96S. aureus isolates from invasive infections were collected. Among these 96 isolates, 55 (56.3%) were identified as MRSA through the cefoxitin disc diffusion test and tested positive for mecA. The sources of invasive S. aureus infections included bacteremia (53.1%), cellulitis (16.7%), pneumonia (9.4%), osteomyelitis (6.3%), endocarditis (6.3%), peritonitis (5.2%), and arthritis (4.3%). Information regarding the isolates of MRSA, MLST, spa types, and SCCmec types was presented in Table 2. All tested S. aureus isolates exhibited genetic diversity. We identified twelve distinct sequence types within the 96 isolates, with ST59 being the most frequently represented clone (25.0%, 24/96), followed by ST7 (16.7%, 16/96), ST188 (14.6%, 14/96), ST398 (11.5%, 11/96), ST5 (0.9%, 9/96), and ST239 (0.6%, 6/96). Notably, ST59 and ST7 were the dominant sequence types for MRSA and MSSA, respectively. Each of the remaining sequence types had no more than five isolates.
 |
Table 2 Molecular Characteristics of Invasive S. aureus Isolates |
The spa typing analysis discriminated the 96S. aureus isolates into 25 spa types, which were further clustered into 9 CCs (clonal complexes). Among these, spa t437 was the most common type (15.6%, 15/96), followed by t189 (14.6%, 14/96), t091 (12.5%, 12/96), t9353 (0.7%, 7/96), and t1250 (0.6%, 6/96). Each of the remaining spa types had no more than five isolates. Using SCCmec typing, we identified four types (types II, III, IV, and V) among the 55 MRSA isolates. Type IV was the most predominant (60.0%, 33/55), followed by type III (18.2%, 10/55), type II (12.7%, 7/55), and type V (9.1%, 5/55). In contrast to previous reports on Chinese isolates, where t30 and t37 were the most common spa types typically associated with ST239,16,17 our study revealed a significantly lower proportion of t37 and t30, as well as ST239 and ST5. Generally, our data demonstrates that invasive S. aureus isolates exhibit genetic diversity, and the major clones associated with invasive S. aureus infections are undergoing rapid changes in our institution.
Antimicrobial Resistance Profiles of Different S. aureus Genotypes
Table 3 presents the resistance profiles of 96S. aureus isolates to various antimicrobial agents. Notably, all isolates demonstrated susceptibility to both vancomycin and linezolid. Among the 96S. aureus isolates, 66.7% (64/96) demonstrated resistance to three or more classes of tested antimicrobial agents, categorizing them as multidrug-resistant isolates. This category encompassed 50 (52.1%) MRSA and 14 (14.6%) MSSA isolates. The resistance rates among the 96S. aureus isolates exhibited significant variations. Resistance to penicillin, oxacillin, clindamycin, and erythromycin exceeded 50%, while resistance to gentamicin, tetracycline, and rifampicin was less than 30%. Within the MRSA isolates, 50 (52.1%) exhibited resistance to three or more antibiotics, 43 (44.8%) displayed resistance to four or more antibiotics, and 28 (29.2%) were resistant to five or more antibiotics. Among the MSSA isolates, 14 (14.6%) were resistant to three or more antibiotics, 8 (8.3%) demonstrated resistance to four or more antibiotics, and 3 (3.1%) displayed resistance to five or more antibiotics. The resistance profiles of S. aureus isolates were found to be genotype-dependent. Notably, ST239 isolates displayed the multidrug-resistant phenotype, with high resistance levels (≥ 80%) observed against all tested antibiotics. ST59 isolates exhibited a higher resistance rate to penicillin (100.0%), oxacillin (100.0%), clindamycin (70.8%), and erythromycin (70.8%). In contrast, ST7, ST188, and ST398 isolates were more resistant to penicillin (100%, 78.6%, and 81.8%, respectively) than to other antibiotics.
 |
Table 3 Antimicrobial Resistance Profiles of S. aureus Causing Invasive Infections Arranged by STs |
Relationship Between Biofilm Production and Methicillin Resistance, MLST, and spa Type
Biofilm formation was observed in 88.5% (85/96) of the isolates (Figure 1A). Among 96 invasive S. aureus isolates, weak biofilm formation was observed in 62.5% (60/96) of isolates, while strong biofilm formation was evident in 26.0% (25/96) of isolates. Additionally, eleven isolates were classified as biofilm formation-negative isolates (non-adherent). Within the group displaying strong biofilm formation (Figure 1A), the majority of isolates (56.0%, 14/25) belonged to the ST7. The assay exhibited excellent reproducibility.
Previous studies have suggested that isolates carrying the mecA gene tend to exhibit stronger biofilm-forming ability compared to mecA-negative isolates.18 However, in our current study, we observed that MSSA isolates demonstrated higher biofilm-forming ability than MRSA isolates (1.08 ± 0.80 vs 0.60 ± 0.40, P = 0.0002) (Figure 1B). The reason for this inconsistency may stem be the variations in the sources of the isolates, encompassing distinct clones of S. aureus. To further investigate whether specific genetic lineages contribute to the enhanced biofilm-forming ability of MSSA, we conducted a comparative analysis of biofilm formation across different sequence types and spa types. Our analysis revealed significant variations in biofilm formation among different sequence types (Figure 1C). Notably, ST7 isolates exhibited markedly stronger biofilm formation than ST59 (1.91 ± 0.65 vs 0.63 ± 0.37, P < 0.0001), ST188 (1.91 ± 0.65 vs 0.63 ± 0.25, P < 0.0001), ST398 (1.91 ± 0.65 vs 0.30 ± 0.19, P < 0.0001), ST5 (1.91 ± 0.65 vs 0.30 ± 0.19, P < 0.0001), and ST239 (1.91 ± 0.65 vs 0.55 ± 0.37, P = 0.0001) isolates. Additionally, ST188 isolates exhibited stronger biofilm-forming ability than ST398 (0.63 ± 0.25 vs 0.30 ± 0.12, P = 0.0006) and ST5 (0.63 ± 0.25 vs 0.30 ± 0.19, P = 0.0034) isolates. Regarding spa types, t091 isolates demonstrated significantly greater biofilm production compared to t437 (1.96 ± 0.64 vs 0.59 ± 0.36, P < 0.0001), t189 (1.96 ± 0.64 vs 0.77 ± 0.41, P < 0.0001), t9353 (1.96 ± 0.64 vs 0.43 ± 0.27, P < 0.0001), t1250 (1.96 ± 0.64 vs 0.33 ± 0.12, P < 0.0001) and other spa types (1.96 ± 0.64 vs 0.69 ± 0.51, P < 0.0001) (Figure 1D).
Distribution of Biofilm-Related Genes
In previous studies, there has been ongoing debate over whether biofilm-related genes can be considered indicators of biofilm-forming ability in S. aureus strains.19–21 To investigate the potential association between the biofilm-forming ability of invasive S. aureus isolates and biofilm-related genes, we selected prominent sequence types of S. aureus prevalent in our institution, specifically ST59 (24 isolates), ST7 (16 isolates), ST188 (14 isolates), and ST398 (11 isolates), as experimental strains. We conducted tests to determine the presence of ten biofilm-related genes. Table 4 presents the prevalence of biofilm-associated genes among isolates of different sequence types. It is worth noting that all isolates tested positive for icaA, icaD, and clfA. However, the prevalence rates of fnbB and sdrD in ST59 isolates were notably low, with carrying rates of 12.5% and 20.8%, respectively. In contrast, among ST7 isolates, the prevalence rates for fnbA, fnbB, clfB, sdrC, and sdrE ranged from 50.0% to 93.8%. Furthermore, sdrD existed in all ST7 isolates. Among ST188 isolates, the carrying rates for fnbA, fnbB, sdrC, and sdrD were relatively low, ranging from 14.3% to 78.6%. All ST398 isolates did not carry fnbA, and the carrying rates for sdrC and sdrD were 9.1%. These findings suggest variability in the presence of biofilm-related genes among different sequence types of S. aureus isolates.
 |
Table 4 Analysis of Biofilm-Related Genes Carrying of the Main Clones |
Expression Levels of icaA Involved in Biofilm Formation Using RT-qPCR
To investigate the factors contributing to the strong biofilm formation observed in ST7 isolates, we randomly selected six isolates from the four major sequence types (ST7, ST59, ST188, and ST398). Remarkably, during our analysis of gene expression levels, we observed that ST7 isolates exhibited significantly higher expression levels of icaA compared to ST59 (P = 0.0012), ST188 (P = 0.0107), and ST398 (P = 0.0002) isolates (Figure 2). These findings align with the robust biofilm-forming characteristics observed in ST7 isolates. This suggests that, while the prevalence rates of biofilm-related genes may not differ significantly, their expression levels can have a substantial impact on biofilm formation. This implies that the high expression of the icaA gene may be the reason behind the strong biofilm-forming ability observed in ST7 isolates.
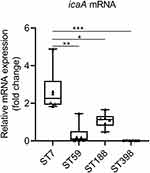 |
Figure 2 The expression levels of icaA. S. aureus N315 was used as a control (relative expression = 1), and gyrB was used as the internal reference gene. *P < 0.05; **P < 0.01; ***P < 0.001. |
Discussion
This study investigated the molecular characteristics, antibiotic susceptibility, and biofilm-forming capabilities of S. aureus isolates recovered from invasive infections in a tertiary hospital in China from 2020 to 2022. We compared MRSA and MSSA isolates and analyzed differences in biofilm production among different sequence types. The key objective of this study was to elucidate the genetic diversity of invasive S. aureus strains to provide insights that can inform infection control efforts and antibiotic stewardship interventions.
MLST and spa typing revealed that ST59-t437 was the most prevalent MRSA isolates, while ST7-t091 predominated among MSSA isolates. A major finding was the emergence of ST7 as the second most common sequence type among invasive S. aureus strains in this setting, closely following endemic ST59. ST7 merits attention as an important epidemic S. aureus clone circulating in China, typically associated with agricultural sources such as pig farms, soil, and ground surfaces.22 Moreover, it is a major genotype of S. aureus found in pork (with a detection rate of 57.5%) and a common cause of bovine mastitis.23,24 This lineage is also prevalent in various food sources.25 Clinically, the ST7 clone often causes skin and soft tissue infections, bacteremia, pneumonia, and musculoskeletal infections in humans, especially among children.26–28 In China specifically, ST7 has been reported as the predominant S. aureus clone causing skin infections in Wenzhou29 and was identified as the second most common lineage among isolates during the COVID-19 pandemic in Wuhan.30 Recently, an foodborne outbreak attributed to ST7 also occurred in Hainan.31 Taken together, these prior reports and our current findings showing the rising prevalence of ST7 in invasive isolates are concerning and suggest this clone may become dominant in China in the coming years. Continued monitoring of the ST7 lineage is warranted concern given its increasing clinical significance regionally.
The ability of pathogenic bacteria to form biofilms significantly contributes to antibiotic resistance and the persistence of chronic infection by aiding in evading host immunity.32 Biofilm formation has been closely associated with persistent staphylococcal infections.33 Bacteria forming biofilms can evade the host immune system.34 In this study, we assessed biofilm production and the carriage of associated genes across isolates. Although the biofilm-associated icaA gene was ubiquitous (Table 4), ST7 isolates exhibited markedly greater biofilm formation phenotypes compared to other lineages (Figure 1C). This discordance between biofilm-related genetics and phenotypes suggests that factors beyond gene presence, such as regulatory and expression differences, may account for enhanced biofilm production in certain clones like ST7.
Staphylococcal biofilm formation is largely attributed to the polysaccharide intercellular adhesin (PIA) encoded by the ica operon.35,36 In MRSA, surface proteins inhibited by agr suppress biofilm production, while in MSSA, PIA generated via the ica locus plays a major role.37,38 Considering that the majority (93.8%) of ST7 isolates in this study were MSSA, we examined icaA expression to investigate the mechanisms underlying this clone’s robust biofilm phenotypes. We found significantly higher icaA expression in ST7 compared to other lineages, aligning with its enhanced biofilm production. These findings suggest a lineage-specific regulation of biofilm formation in ST7, whereby this emerging clone may utilize ica-mediated PIA production as a key virulence strategy. The ability to form substantial biofilms may facilitate ST7’s persistence and increasing prevalence regionally. Further genetic analysis is warranted to elucidate the molecular factors driving ST7’s proficient biofilm formation.
Antibiotic resistance is a major concern with S. aureus. In this study, we found genotype-dependent variability in resistance profiles of invasive isolates. Most MRSA isolates (90.9%) displayed multidrug resistance to at least three tested antibiotics. ST239 MRSA stood out with significantly higher levels of multidrug resistance versus other sequence types. Interestingly, however, despite its robust biofilm production, ST7 did not exhibit broader antibiotic resistance than other lineages. Other resistance determinants, such as the acquisition of specific antibiotic resistance genes, likely play a more important role in defining the antibiotic susceptibility of ST7 clone. Further investigation is needed to elucidate the differential resistance mechanisms among predominant sequence types.
This study has noteworthy limitations. Firstly, the sample size was relatively small, potentially limiting the generalizability of the findings to the broader ST7 genotype population. Additionally, a more in-depth genomic analysis is required to elucidate the underlying mechanisms contributing to the strong biofilm formation observed among ST7 isolates. To advance this research, future studies should employ larger sample sizes and conduct comparative genomics to gain a more comprehensive understanding of genetic factors influencing virulence in this lineage. Expanding both the sampling and genomic characterization will fortify the conclusions and address the current study’s limitations.
In conclusion, this study identified that invasive S. aureus isolates were predominantly of the ST59-t437 and ST7-t091 lineages. Molecular characteristics, antibiotic susceptibility patterns, and biofilm formation abilities differed across sequence types. Notably, our research revealed an association between specific S. aureus genetic lineages and enhanced biofilm formation in invasive isolates. The emergence of the ST7 clone with robust biofilm-producing capabilities could significantly complicate treatment approaches. Overall, this study provides valuable insights into the molecular characteristics of invasive S. aureus isolates in this region. Continuous surveillance of emerging successful lineages is critical to help guide infection control strategies against invasive S. aureus infections.
Abbreviations
MLST, Multi-locus Sequence Typing; SCCmec, chromosomal cassette mec; MRSA, Methicillin-resistant S. aureus; MSSA, Methicillin-sensitive S. aureus; CLSI, Clinical and Laboratory Standards Institute; ST, sequence type; NT, non-typeable; TSB, Tryptic Soy Broth; RT-qPCR, Real-time quantitative PCR; PIA, polysaccharide intercellular adhesin; ica, intercellular adhesion.
Ethics Statement
Ethical approval for this study was obtained from the Ethical Committee of the First Hospital of Nanchang, Jiangxi Province, China. Informed patient consent was waived due to the nature of S. aureus clinical isolation, which is an integral part of routine hospital laboratory procedures. We have ascertained that the isolated samples contain no identifiable patient data.
Funding
This work was supported by the Science and Technology Project of Jiangxi Health Commission (No.202140032).
Disclosure
The authors declare no conflicts of interest.
References
1. Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi:10.1128/CMR.00134-14
2. Cosgrove SE, Fowler VG. Management of methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2008;46(Suppl 5):S386–S393. doi:10.1086/533595
3. Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi:10.3201/eid0809.020063
4. Khatoon Z, McTiernan CD, Suuronen EJ, Mah T-F, Alarcon EI. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4:
5. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi:10.1126/science.284.5418.1318
6. Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–7121. doi:10.1128/AEM.02636-13
7. Arciola CR, Campoccia D, Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16:397–409. doi:10.1038/s41579-018-0019-y
8. David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–687. doi:10.1128/CMR.00081-09
9. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi:10.1128/JCM.38.3.1008-1015.2000
10. Koreen L, Ramaswamy SV, Graviss EA, et al. Spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42(2):792–799. doi:10.1128/JCM.42.2.792-799.2004
11. Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41(12):5442–5448. doi:10.1128/JCM.41.12.5442-5448.2003
12. Zhang K, McClure J-A, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi:10.1128/JCM.43.10.5026-5033.2005
13. Kondo Y, Ito T, Ma XX, et al. Combination of multiplex PCRs for Staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in Junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi:10.1128/AAC.00165-06
14. Xiao Y, Han W, Wang B, et al. Phylogenetic analysis and virulence characteristics of methicillin-resistant Staphylococcus aureus ST764-SCC mec II: an emerging hypervirulent clone ST764-t1084 in China. Emerg Microbes Infect. 2023;12(1):2165969. doi:10.1080/22221751.2023.2165969
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
16. Liu Y, Wang H, Du N, et al. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother. 2009;53(2):512–518. doi:10.1128/AAC.00804-08
17. Chen H, Liu Y, Jiang X, Chen M, Wang H. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob Agents Chemother. 2010;54:1842–1847. doi:10.1128/AAC.01563-09
18. Leshem T, Schnall B-S, Azrad M, et al. Incidence of biofilm formation among MRSA and MSSA clinical isolates from hospitalized patients in Israel. J Appl Microbiol. 2022;133(2):922–929. doi:10.1111/jam.15612
19. Bimanand L, Taherikalani M, Jalilian FA, et al. Association between biofilm production, adhesion genes and drugs resistance in different SCCmec types of methicillin resistant Staphylococcus aureus strains isolated from several major hospitals of Iran. Iran J Basic Med Sci. 2018;21(4):400–403. doi:10.22038/IJBMS.2018.19378.5132
20. Ohadian Moghadam S, Pourmand MR, Aminharati F. Biofilm formation and antimicrobial resistance in methicillin-resistant Staphylococcus aureus isolated from burn patients, Iran. J Infect Dev Ctries. 2014;8:1511–1517. doi:10.3855/jidc.5514
21. Puah SM, Tan JAMA, Chew CH, Chua KH. Diverse profiles of biofilm and adhesion genes in Staphylococcus aureus food strains isolated from sushi and sashimi. J Food Sci. 2018;83:2337–2342. doi:10.1111/1750-3841.14300
22. Yan X, Yu X, Tao X, et al. Staphylococcus aureus ST398 from slaughter pigs in northeast China. Int J Med Microbiol. 2014;304:379–383. doi:10.1016/j.ijmm.2013.12.003.
23. Zhu Z, Liu X, Chen X, et al. Prevalence and virulence determinants of Staphylococcus aureus in wholesale and retail pork in Wuhan, Central China. Foods Basel Switz. 2022;11:
24. Zhou Y, Li X, Yan H. Genotypic characteristics and correlation of epidemiology of Staphylococcus aureus in healthy pigs, diseased pigs, and environment. Antibiotics. 2020;9:
25. Liao F, Gu W, Yang Z, et al. Molecular characteristics of Staphylococcus aureus isolates from food surveillance in southwest China. BMC Microbiol. 2018;18(1):91. doi:10.1186/s12866-018-1239-z
26. Gu -F-F, Chen Y, Dong D-P, et al. Molecular epidemiology of Staphylococcus aureus among patients with skin and soft tissue infections in two Chinese hospitals. Chin Med J. 2016;129(19):2319–2324. doi:10.4103/0366-6999.190673
27. Li S, Sun S, Yang C, et al. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Front Microbiol. 2018;9:332. doi:10.3389/fmicb.2018.00332
28. Wu S, Huang J, Wu Q, et al. Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front Microbiol. 2018;9:2767. doi:10.3389/fmicb.2018.02767
29. Yu F, Liu Y, Lv J, et al. Antimicrobial susceptibility, virulence determinant carriage and molecular characteristics of Staphylococcus aureus isolates associated with skin and soft tissue infections. Braz J Infect Dis. 2015;19:614–622. doi:10.1016/j.bjid.2015.08.006.
30. Gu J, Shen S, Xiong M, et al. ST7 becomes one of the most common Staphylococcus aureus clones after the COVID-19 epidemic in the city of Wuhan, China. Infect Drug Resist. 2023;16:843–852. doi:10.2147/IDR.S401069
31. Guo Y, Yu X, Wang J, et al. A food poisoning caused by ST7 Staphylococcal aureus harboring sea gene in Hainan province, China. Front Microbiol. 2023;14:1110720. doi:10.3389/fmicb.2023.1110720
32. Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;121(s136):1–51. doi:10.1111/apm.12099
33. Mohamed JA, Huang DB, Jiang Z-D, et al. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol. 2007;45(1):121–126. doi:10.1128/JCM.01128-06
34. Roilides E, Simitsopoulou M, Katragkou A, Walsh TJ. How biofilms evade host defenses. Microbiol Spectr. 2015;3:3. doi:10.1128/microbiolspec.mb-0012-2014
35. Mack D, Fischer W, Krokotsch A, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi:10.1128/jb.178.1.175-183.1996
36. Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F, Kaufmann SHE. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–5433. doi:10.1128/IAI.67.10.5427-5433.1999
37. O’Neill E, Pozzi C, Houston P, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45(5):1379–1388. doi:10.1128/JCM.02280-06
38. Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi:10.1128/AEM.02073-07
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

