Back to Archived Journals » Nanobiosensors in Disease Diagnosis » Volume 4
Molecular biosensors: promising new tools for early detection of cancer
Authors Altintas Z, Tothill I
Received 15 July 2014
Accepted for publication 30 August 2014
Published 19 January 2015 Volume 2015:4 Pages 1—10
DOI https://doi.org/10.2147/NDD.S56772
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Chulhee Choi
Zeynep Altintas, Ibtisam E Tothill
Biomedical Engineering, Cranfield University, Cranfield, UK
Abstract: Over the last decade, an increasing number of researchers have focused on developing rapid techniques based on biosensor technology for the detection of various human health-related conditions. Cardiovascular disease, cancer, diabetes, and infectious diseases are at the top of these investigations. Huge advances have taken place in a wide range of disciplines, with improved sensitivity, specificity, and reproducibility of the designed methodologies. Advances in other scientific disciplines, including genetics, chemical and microelectronic engineering, computational sciences, and medicine have also impacted on the developments taking place in the diagnostics field. As a result of these developments, more stable, rapid, and reliable sensing platforms have been successfully constructed for specific and ultrasensitive biomarker-based disease diagnostics. These achievements were also a result of improving sensor production quality, developing appropriate surface chemistries, selection of high affinity ligands for target biomarkers, and using nanomaterials such as carbon nanotubes, magnetic/gold nanoparticles, or quantum dots for signal amplification. Moreover, use of bioinformatics tools, combined with proteomics and genomics technologies and metabolomics, has significantly improved the knowledge and discovery of new biomarkers that can be used for early detection. Due to these significant outputs, coupled with advances in miniaturization and nanotechnology, the demand for development and production of sensor systems, including optical, piezoelectric/acoustic, electrochemical, and colorimetric, has gradually increased during the last decade. Moreover, not only protein and gene markers, but also secondary metabolites and cell signaling pathway molecules can be used for disease detection. To date, diagnostic tests for the early detection of cancer biomarkers are available on the market, and these provide noninvasive/minimally invasive tests that can play a crucial role both in successful treatment and patient survival. This paper reviews disease biomarkers, post-genomic technology, and use of molecular biosensors for early detection of cancer.
Keywords: cancer, biomarker discovery, post-genomic technology, molecular biosensors, disease diagnostics
Introduction
Cancer is a multifactorial molecular disease that includes multistage development of tumor cells. Cancer can be caused by a range of genetic or environmental factors, such as exposure to carcinogenic chemicals or radiation, or have a microbiological cause, including bacterial (eg, stomach cancer) or viral (eg, cervical cancer) infection. The characteristic behavior of the cancer is a process reflecting the complexity of malignant disease. This process includes promoter proliferative signaling, escape from growth suppressors, resistive cell death, replicative immortality, triggering of angiogenesis, and activation of invasion and metastasis. Genome instability, inflammation, and epigenetic changes are the underlying reasons for these. Despite the common features of neoplasms, there are a variety of types of cancer due to genetic diversity. Therefore, profiling each type of cancer is important to be able to develop an efficient and specific treatment without causing any damage to other tissues and organs.1
Determination of the responsible genes and protein biomarkers as early as possible before development of tumors is key to developing methods for early detection of cancer. Molecular biology and genetics play an important role in mapping the relevant molecular biomarkers with the combination of post-genomics technologies.2–4 Bioinformatics applications for patient profiling can provide the most significant information using sequencing techniques, making it possible to use these molecules not only at the molecular biology level but also for early diagnosis. Both genes and proteins, abnormalities of which lead to cancer, can be used as indicators of specific types of cancer. These indicators are known as biomarkers and can exist in tumors and body fluids, such as blood, urine, and sputum.5 Due to the invasive nature of a biopsy, and the disadvantages of currently available diagnostic tools6–17 (Table 1), the possibility of using body fluids for the diagnosis of cancer has emerged, and this opens up enormous opportunities for biosensor technology. These methods should provide information to assist clinicians in making successful treatment decisions and increasing patient survival.
  | Table 1 Currently available diagnostic tools for cancer with their advantages and disadvantages |
The major applications of nanobiosensors in molecular diagnostics cover nanotechnology-based biochips and microarrays, nanotechnology-based cytogenetics, application of nanoparticles to track stem cells, identification of cancer cells or biomolecules, and biomarker discovery.18 Biological assays that measure the activity or presence of selected compounds through the use of microfluidics/lab-on-a-chip systems can provide more sensitive, reliable, and rapid results when certain nanomaterials are used as labels or tags.19–21
Advances in the development of molecular biosensors and biomarker discovery combined with post-genomics technology have provided a deeper understanding of cancer, with better biomarkers to use for its detection. Hence, improved diagnostics platforms have emerged that are able to detect these biomarkers and also employ nanomaterials for enhanced performance. These fascinating improvements can be employed for detecting cancer at an earlier stage, thereby helping in devising a more effective treatment and decreasing mortality rates from cancer, as well as cost.
Disease biomarkers and post-genomic technology
Cancer markers
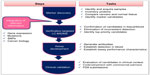
Malignant cells produce biological substances due to progression of disease and also because of other factors such as inflammation. These substances can be used to detect the cancer and are called biomarkers. The level of biomarkers can be quantified, higher or lower in cancer cases than normal levels in body fluids and/or tissues, using a number of techniques (Figure 1). Intracellular or intercellular compounds, such as hormones, mucin, isoenzymes, oncogenes, oncofetal antigens, tissue-specific proteins, circulating ribonucleic acids, and some glycoproteins and glycolipids, may be used as indicators of cancer.22–24 These molecules can be used for development of therapy, determination of existing cancer type, or estimation of possible future relapse. Cancer has many subgroups depending on the organ involved, including prostate, lung, colon, breast, ovarian, brain, and others, and each type has its own associated biomarkers.25 Some of the biomarkers are organ-specific, whereas others are common to other types of cancer. For example, neuron-specific enolase and a breast cancer gene are specific for lung and breast carcinomas, respectively; whereas cancer antigen 15-3, cancer antigen-125, and carcinoembryonic antigen may indicate breast, lung, or colon cancer, respectively. Further, biomarkers can show different concentration level in different type of cancer. Information regarding the type of biomarker linked to the type of cancer and their concentration level is given in the literature.22,25
  | Figure 1 Techniques used for biomarker identification and detection. |
Due to the importance of cancer marker specificity and sensitivity for the target organ, multiple biomarker assays need to be performed in order to increase the accuracy of cancer detection.26,27 Gene biomarkers can show more specificity for each cancer type because of genome diversity, which leads to the occurrence of mutations in different locations of a particular gene. A variety of genes has been linked to somatic mutations in human tumor cells or tissues, and more are being discovered every year. These mutated genes include oncogenes and tumor suppressor genes, as well as genes encoding proteins that perform key functions in regulation of the cell cycle, DNA repair, and enzyme activity. For example, P53 gene mutations constitute the most critical cancer mutation due to the central role of this gene in cell division and cell death mechanisms. It is called the guardian of the genome, regulating the cell cycle and functioning as a tumor suppressor. P53 is located on the short arm of chromosome 17 in the human genome.28 Although there are some common mutations, such as substitution of an arginine for a proline in the codon 72 position, other cancer-specific mutations are also observed in other genes. Moreover, a mutation occurring in the same location can initiate different cancers depending on the geographical location, ethnic background, and/or gender of the patient. Sonoyama et al investigated P53 proline mutation and found that it caused pancreatic cancer in males,29 whereas the same mutation decreased the risk of breast cancer in Arab women.30
Gene markers can assist in the determination of subtypes of cancer. For example, lung cancer has two main subgroups that show completely different developmental stages during the disease. Non-small cell lung cancer is affected by codon 273 mutation, whereas small cell lung cancer shows a significant relationship with a mutation of codon 248 in the P53 gene.31 Mutations in responsible genes can be used as biomarkers for determination of cancer cases and development of therapy, and so can their protein products. The epidermal growth factor receptor is responsible for tumor invasion, angiogenesis, cell proliferation, and apoptosis.32 A mutated version of this protein is common in lung cancer, and epidermal growth factor receptor inhibitors can be used as treatment. Precise determination and mapping of gene and protein markers for each type of cancer is essential.33 Advances in genomic and post-genomic technology have been changing the landscape day by day in a positive way, and next generation sequencing techniques offer an opportunity to improve both biomarker discovery and the number of biomarkers that can be used for early and precise detection of cancer.34,35
Post-genomic technology for discovery of biomarkers
Early diagnosis of cancer is crucial for patient survival and successful prognosis of the disease, so sensitive and specific methods are required for its detection. Analysis of biomarkers in body fluids is one of the methods used for detection of the disease. Hence, multi-marker profiles (presence and concentration) are essential for the early diagnosis of cancer. Nowadays, biomarkers have applications in diagnosis, cancer staging, prognosis, monitoring for disease progression, and also in development of therapy. The combination of different scientific fields and technologies for biomarker discovery are momentous for their precise identification, characterization, and verification.36 The development of proteomic methods to find new and novel biomarkers is closely related to the epochal efforts of genomic investigations.
The Human Genome Project and the knowledge of other genomes have led to the construction of protein databases. In addition to proteomic information, which was built based on genomic outputs, gene expression data identified by sequencing techniques or microarrays have redounded further knowledge to be used in cancer cases.37,38 Genomic and proteomic analyses have been performed to determine potential biomarkers, since the combined data are much more reliable and precise than the data obtained using a single methodology. Therefore, integration of the techniques and investigations gives more detailed insights into the identification and verification of a potential cancer biomarker.37,39 Moreover, the knowledge obtained may result in finding more specific gene or protein biomarkers for a particular cancer type, that have a critical role in early determination and diagnosis as well as in prognosis and disease follow-up.40–42
Protein chips can also be produced and used for biomarker discovery. Here, complementary DNA-encoding tagged proteins are expressed, and the proteins are isolated and printed on a slide surface. The slides are then investigated for protein–protein interactions, determination of antibodies, and protein profiling. The protein arrays lead to detection of possible novel biomarkers and enable a deeper understanding of the printed proteins related to signaling pathways on the slides.43,44
There are several stages between the discovery of biomarkers and their clinical use in patients with cancer.45 Post-genomic technologies and bioinformatics tools help to identify novel candidate markers with the integration of relevant information, such as gene expression, mutations, single nucleotide polymorphisms, and cancer biology. Verification of candidate biomarkers is then accomplished by liquid chromatography-mass spectrometry to identify the top priority candidates and eliminate inconsistent ones. Selected candidates need to be screened and investigated in blood samples using the developed bioassays for characterization of all required parameters prior to clinical validation.46 Research is ongoing to discover new biomarkers that are more specific and sensitive for each type of cancer using the post-genomic plethora of data available from the Human Genome Project.47 Figure 2 represents all stages from discovery to clinical use of cancer biomarkers, along with the tasks required for each.
Molecular biosensors for early detection of cancer
Interest in biosensor technologies has increased specifically for diagnosis of cancer, and the greatest advantage is noninvasive and effective early detection. A biosensor is an analytical device incorporating a molecular recognition receptor associated or integrated with a physiochemical transducer (Figure 3).25 Biosensors can be classified as point-of-care devices that enable analysis of clinical samples in the home or at the doctor’s surgery. In order to develop an appropriate biosensor strategy to detect cancer, a specific biomarker or an array of biomarkers need to be identified to ensure specificity of the devices. To detect a specific biomarker, optimal recognition materials need to be used for the receptor molecule in the biosensor design. A range of molecular recognition materials has been used in the development of biosensors, including natural materials such as antibodies, or synthetically generated (artificial) molecular imprinting polymers, aptamers, and phage display peptides.25 A transducer is a device that converts recognition signal events into electrical (often digital) signals, and can be electrochemical, optical, calorimetric, piezoelectric/acoustic, or magnetic.
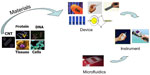  | Figure 3 Construction of a biosensor: from materials, to devices, to systems. |
A vast number of research papers have been published in the literature on the development of biosensors to detect cancer.48–57 Biosensors have the potential to detect an array of biomarkers specifically and at the low level of marker concentration existing in the early stages of the disease. Different systems have been developed based on a range of transducers, such as optical-based surface plasmon resonance sensors, piezoelectric-based quartz crystal microbalance sensors, and electrochemical-based sensors (Table 2). Advances in nanotechnology have had a huge impact on improving current molecular diagnostic techniques, such as the development of point-of-care tests and personalized medicine, as well as the combination of diagnostics with therapeutics. These also include major applications in silicon biochips and microarrays, nanotechnology-based cytogenetic use of nanoparticles to track stem cells, and identification of single cells or molecules.18
An electrochemical molecular beacon biosensor has been developed for sequence-specific recognition of DNA, which is important for cancer diagnostics and gene therapy. A sequence-specific DNA biosensor was constructed in one study using a ferrocene redox probe modified molecular beacon. Double-stranded DNA can be quantified using this biosensor in the concentration range of 350 pM to 25 nM, with a detection limit of 275 pM.58 Further, Xuan et al used molecular beacon-based DNA detection with exonuclease III-assisted target recycling as a signal amplification method. The sensor was able to successfully detect DNA in homogeneous solution, and appears to be a promising technology for integration of the technique into a portable, cost-effective DNA sensing device.59 These investigations indicate great potential for detection of cancer using genetic biomarkers with extreme sensitivity and selectivity.60 Molecular beacon-based biosensors can also be used for the detection of protein biomarkers, small molecules, and RNA.61–63
A potentiometric biosensor combining the effective and specific molecular imprinting technique with a self-assembled monolayer was developed for detection of cancer as a point-of-care diagnostics tool. The sensor was initially tested with carcinoembryonic antigen as a pancreatic biomarker and then used to discriminate between damaged and normal fibrinogen, which is important in the diagnosis of bleeding disorders. The protein structures were modeled using computational simulation to determine the morphological changes and sensitivity of the sensor system to these conformational changes in the protein.64 Mathur et al65 constructed a biosensor using surface imprinting for quantification of pleural mesothelioma cancer biomarkers. Hyaluronan and proteoglycan link protein 1 (HAPLN1) was imprinted in duplicate and detection of spiked HAPLN1 was successful in serum. For this, HAPLN1 was imprinted on a gold electrode with hydroxyl-terminated alkane thiols, which formed a self-assembled monolayer around HAPLN1. The target analyte was then washed away and its imprint was used for detection of HAPLN1 using an electrochemical open-circuit potential method. The selectivity of the imprints was verified by matrix-assisted laser desorption ionization-time of flight mass spectrometry.65 A piezoelectric sensor was developed for label-free, rapid, and cost-effective determination of prostate-specific antigen and α-fetoprotein. A titanate zirconate ceramic resonator was used as the transducer and a dual system was designed to build two ceramic resonators that were connected and parallel to each other. One resonator was used as the sensing unit and the other as the control to minimize temperature fluctuation and to obtain a stable frequency for the measurements. The sensor could successfully detect prostate-specific antigen and α-fetoprotein up to 0.25 ng/mL in a short time (30 minutes) with a small sample volume (1 μL). The results confirmed that this ceramic resonator-based piezoelectric biosensor could be used with different chemical interfaces, and the small size of the resonators provides an opportunity to fabricate sensor arrays for multiplex detection, which is crucial in the precise diagnosis of cancer cases.66
Furthermore, bioassays measuring the activity or presence of target cancer biomarker molecules through molecular sensing systems enable more sensitive and rapid detection when nanomaterials are used as signal amplifiers or tags.48 Magnetic-based immunoassay techniques have been developed in which target analyte molecules or microorganisms can be labeled with magnetic nanoparticles via an appropriate antibody. Short fragments of DNA can also be tagged with gold nanoparticles for the detection of targeted gene sequences. Multicolor optical coding using quantum dots is another important application in molecular diagnostics and is based on embedding of different-sized quantum dots into polymeric microbeads.22
Although various nanoparticles have been used in molecular diagnostics, the most commonly used ones are magnetic nanoparticles, gold nanoparticles, and quantum dots. Nanoparticles have a wide range of applications for detection of different types of disease, such as cancer, infectious diseases, and neurological disorders.67 For example, quantum dots and magnetic iron oxide nanoparticles can be designed for dual-mode imaging of cancer in which the best characteristics of each are combined to develop a single nanoparticle probe that can produce clinically useful images of cancer tumors and/or molecules. Silica nanoparticles 30 nm in size incubated with rhodamine (a bright fluorescent dye) and iron oxide nanoparticles 9 nm in size produced nanoparticles 45 nm in diameter that performed better on fluorescent imaging and magnetic resonance imaging (MRI) than the individual particles. These particles were also used to visualize lung tumor cells by using an anti-lung cancer cell antibody attached to the particles. When the antibodies recognize the lung cancer cells and attach to them, fluorescence microscopy can be used for their detection.68
Bioconjugated quantum dots are important features of molecular diagnostics for multiple profiling of biomarkers, correlation with disease progression, and response to therapy.69 Such applications improve the ability to estimate the outcome of drug therapy in disease management. The success of these techniques can be increased using bioinformatics and systems biology to link each patient’s molecular profile with disease diagnostics and treatment selection. This approach was applied for simultaneous determination of multiple biomarkers in prostate cancer.70 El-Sayed et al used gold nanoparticle-conjugated anti-epidermal growth factor receptor antibodies that bind to the surface of cancer cells specifically and homogenously. The binding affinity was found to be 600% higher for cancerous cells than for noncancerous cells. The same group also used gold nanoparticles to detect cancer cells noninvasively and destroy them using selective laser photothermal therapy.71
Manganese oxide nanoparticles were combined with MRI for visualization of the anatomic structure of the mouse brain. The images produced were as clear as those acquired by histology. This novel MRI technique may improve our ability to diagnose neurological disorders such as Parkinson’s disease, stroke, and Alzheimer’s disease.72 Moreover, affinity ligands such as antibodies can be attached to these nanoparticles, and distinguish and selectively bind to receptors on the cancer cell surface in the mouse brain due to metastases. This method can also be developed for other cancer types using appropriate antibodies. Infectious diseases affect the immune system and lower the tolerance of the human body to other health problems.73 The pathogens may initiate a response in the cell and are identified by the receptors responsible for the cancer. Therefore, detection of these microorganisms is also critical in some cancer cases, and application of nanomaterials in molecular diagnostics allows rapid and sensitive detection of these pathogens. It is possible to detect a single bacterium using bioconjugated nanoparticle-based assays. For this, quantum dots functionalized with multicolor oligonucleotides are used as nanoprobes and a hybridization-based detection method is applied in the presence of different target sequences, creating visible sequence-specific spectral codings.73 Spectroscopic assays combined with silver nanorods have been developed for rapid quantification and characterization of viruses with high specificity and sensitivity and without the need for viral manipulation. This method can recognize spectral differences between viral strains, viruses, and viruses with gene deletions in biological sample.74 Table 3 lists some examples of the use of nanomaterials for detection of cancer.
Conclusion
Diagnosis of cancer using molecular biosensors is an emerging field with a critical role in the construction of highly sensitive, rapid, easy-to-use, and reliable techniques for early and precise detection of cancer. Given that it is crucial to diagnose cancer early to allow successful treatment and recovery in patients, it is very important to have cost-effective devices that can detect multiple cancer biomarkers existing at low concentrations in biological fluids. Surface imprinting techniques, the use of artificial receptors such as aptamers and nanomaterials are promising tools for expanding the current technology of molecular diagnostics and provide point-of-care diagnostic devices, disease care with therapeutics, and personalized medicine. Integration of biomarker discovery into nanodevices with the combination and modification of nanomaterials has improved clinical and research-based applications for cancer in recent years. Further development of these nanosystems and nanoarray devices and use of novel multi-biomarker detection will play a critical role in molecular diagnostics by providing early and rapid diagnosis, decreasing the sample volume required and device costs, and improving the currently available detection methods.
Disclosure
The authors report no conflicts of interest in this work.
References
Weinberg R. The Biology of Cancer. New York, NY, USA: Garland Science; 2013. | |
Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. | |
Chung CC, Ciampa J, Yeager M, et al. Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Hum Mol Genet. 2011;20:2869–2878. | |
Tang W, Fu YP, Figueroa JD, et al. Mapping of the UGT1A locus identifies an uncommon coding variant that affects mRNA expression and protects from bladder cancer. Hum Mol Genet. 2012;21:1918–1930. | |
Altintas Z, Uludag Y, Gurbuz Y, Tothill IE. Surface plasmon resonance based immunosensor for the detection of the cancer biomarker carcinoembryonic antigen. Talanta. 2011;86:377–383. | |
Woolner LB, McDonald JR. Biopsy in cancer diagnosis. Plast Reconstr Surg. 1952;9:388. | |
Kronz JD, Allan CH, Shaikh AA, Epstein JI. Predicting cancer following a diagnosis of high-grade prostatic intraepithelial neoplasia on needle biopsy: data on men with more than one follow-up biopsy. Am J Surg Pathol. 2001;25:1079–1095. | |
Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res. 1994;54:1634–1637. | |
Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. | |
Boice JD Jr, Preston D, Davis FG, Monson RR. Frequent chest X-ray fluoroscopy and breast cancer incidence among tuberculosis patients in Massachusetts. Radiat Res. 1991;125:214–222. | |
de GonzÁlez AB, Darby S. Risk of cancer from diagnostic X-rays: estimates for the UK and 14 other countries. Lancet. 2004;363:345–351. | |
Leach MO, Boggis CR, Dixon AK, et al; MARIBS Study Group. Screening with magnetic resonance imaging and mammography of a UK population at high familial risk of breast cancer: a prospective multicentre cohort study (MARIBS). Lancet. 2005;365:1769–1778. | |
Beets-Tan RG, Beets GL, Vliegen RF, et al. Accuracy of magnetic resonance imaging in prediction of tumour-free resection margin in rectal cancer surgery. Lancet. 2001;357:497–504. | |
Watt I, Stewart I, Anderson D, Bell G, Anderson JR. Laparoscopy, ultrasound and computed tomography in cancer of the oesophagus and gastric cardia: a prospective comparison for detecting intra-abdominal metastases. Br J Surg. 1989;76:1036–1039. | |
Lardinois D, Weder W, Hany TF, et al. Staging of non–small-cell lung cancer with integrated positron-emission tomography and computed tomography. N Engl J Med. 2003;348:2500–2507. | |
Ollinger JM, Fessler JA. Positron-emission tomography. IEEE Signal Process Mag. 1997;14:43–55. | |
Ter-Pogossian MM, Raichle ME, Sobel BE. Positron-emission tomography. Sci Am. 1980;243:170–181. | |
Jain KK. Nanodiagnostics: application of nanotechnology in molecular diagnostics. Expert Rev Mol Diagn. 2003;3:153–161. | |
Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Microfluidic electrochemical immunoarray for ultrasensitive detection of two cancer biomarker proteins in serum. Biosens Bioelectron. 2011;26:4477–4483. | |
Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL, et al. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS). Lab Chip. 2011;11:845–850. | |
Malhotra R, Patel V, Chikkaveeraiah BV, et al. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal Chem. 2012;84:6249–6255. | |
Altintas Z, Tothill I. Biomarkers and biosensors for the early diagnosis of lung cancer. Sens Actuators B Chem. 2013;188:988–998. | |
Altintas Z, Fakanya WM, Tothill IE. Cardiovascular disease detection using bio-sensing techniques. Talanta. 2014;128:177–186. | |
Fakanya WM, Altintas Z, Tothill IE. Biosensors for heart disease diagnosis, In: Biosensors and their application in healthcare. (e-book), Future Science Group, UK; 2013. 128–143. | |
Tothill IE. Biosensors for cancer markers diagnosis. Semin Cell Dev Biol. 2009;20:55–62. | |
Altintas Z, Kallempudi SS, Gurbuz Y. Gold nanoparticle modified capacitive sensor platform for multiple marker detection. Talanta. 2014;118:270–276. | |
Altintas Z, Kallempudi SS, Sezerman U, Gurbuz Y. A novel magnetic particle-modified electrochemical sensor for immunosensor applications. Sens Actuators B Chem. 2012;174:187–194. | |
Isobe M, Emanuel BS, Givol D, Oren M, Croce CM. Localization of gene for human p53 tumour antigen to band 17p13. Nature. 1986;12:84–85. | |
Sonoyama T, Sakai A, Mita Y, et al. TP53 codon 72 polymorphism is associated with pancreatic cancer risk in males, smokers and drinkers. Mol Med Rep. 2011;4:489–495. | |
Alawadi S, Ghabreau L, Alsaleh M, et al. P53 gene polymorphisms and breast cancer risk in Arab women. Med Oncol. 2011;28:709–715. | |
Hainaut P, Pfeifer GP. Patterns of p53 G? T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. | |
Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. | |
Bouchal P, Dvorakova M, Scherl A, Garbis SD, Nenutil R, Vojtesek B. Intact protein profiling in breast cancer biomarker discovery: protein identification issue and the solutions based on 3D protein separation, bottom-up and top-down mass spectrometry. Proteomics. 2013;13:1053–1058. | |
Wheelock CE, Goss VM, Balgoma D, et al. Application of’omics technologies to biomarker discovery in inflammatory lung diseases. Eur Respir J. 2013;42:802–825. | |
Mayr M, Zampetaki A, Willeit P, Willeit J, Kiechl S. MicroRNAs within the continuum of postgenomics biomarker discovery. Arterioscler Thromb Vasc Biol. 2013;33:206–214. | |
Hanash SM, Bobek MP, Rickman DS, et al. Integrating cancer genomics and proteomics in the post-genome era. Proteomics. 2002;2:69–75. | |
Imielinski M, Cha S, Rejtar T, Richardson EA, Karger BL, Sgroi DC. Integrated proteomic, transcriptomic, and biological network analysis of breast carcinoma reveals molecular features of tumorigenesis and clinical relapse. Mol Cell Proteomics. 2012;11:1–7. | |
Peng J, Ritchie JC, Van Meir EG. Proteomics of gliomas: initial biomarker discovery and evolution of technology. Neuro-oncology. 2011;13:926–942. | |
Casado-Vela J, CebriÁn A, Gómez del Pulgar MT, Lacal JC. Approaches for the study of cancer: towards the integration of genomics, proteomics and metabolomics. Clin Transl Oncol. 2011;13:617–628. | |
Chen G, Gharib TG, Huang CC, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. | |
Kanehisa M, Bork P. Bioinformatics in the post-sequence era. Nat Genet. 2003;33:305–310. | |
Ørntoft TF, Thykjaer T, Waldman FM, Wolf H, Celis JE. Genome-wide study of gene copy numbers, transcripts, and protein levels in pairs of non-invasive and invasive human transitional cell carcinomas. Mol Cell Proteomics. 2002;1:37–45. | |
Lueking A, Huber O, Wirths C, et al. Profiling of alopecia areata autoantigens based on protein microarray technology. Mol Cell Proteomics. 2005;4:1382–1390. | |
Pauly F, Smedby KE, Jerkeman M, et al. Identification of B-cell lymphoma subsets by plasma protein profiling using recombinant antibody microarrays. Leuk Res. 2014;38:682–690. | |
Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev. 2010;19:2995–2999. | |
Frangogiannis NG. Biomarkers: hopes and challenges in the path from discovery to clinical practice. Transl Res. 2012;159:197–204. | |
Cantor CR. Orchestrating the Human Genome Project. Science. 1990;248(4951):49–51. | |
Uludag Y, Tothill IE. Cancer biomarker detection in serum samples using surface plasmon resonance and quartz crystal microbalance sensors with nanoparticle signal amplification. Anal Chem. 2012;84:5898–5904. | |
Altintas Z, Tothill IE. DNA-based biosensor platforms for the detection of TP53 mutation. Sens Actuators B Chem. 2012;169:188–194. | |
Wang Y, Zhu X, Wu M, Xia N, Wang J, Zhou F. Simultaneous and label-free determination of wild-type and mutant p53 at a single surface plasmon resonance chip preimmobilized with consensus DNA and monoclonal antibody. Anal Chem. 2009;81:8441–8446. | |
Ladd J, Lu H, Taylor AD, Goodell V, Disis ML, Jiang S. Direct detection of carcinoembryonic antigen autoantibodies in clinical human serum samples using a surface plasmon resonance sensor. Colloids Surf B Biointerfaces. 2009;70:1–6. | |
Lin YY, Wang J, Liu G, Wu H, Wai CM, Lin Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens Bioelectron. 2008;23:1659–1665. | |
Ilyas A, Asghar W, Allen PB, Duhon H, Ellington AD, Iqbal SM. Electrical detection of cancer biomarker using aptamers with nanogap break-junctions. Nanotechnology. 2012;23:1–8. | |
Kallempudi SS, Altintas Z, Niazi JH, Gurbuz Y. A new microfluidics system with a hand-operated, on-chip actuator for immunosensor applications. Sens Actuators B Chem. 2012;163:194–201. | |
Sato Y, Fujimoto K, Kawaguchi H. Detection of a K-ras point mutation employing peptide nucleic acid at the surface of a SPR biosensor. Colloids Surf B Biointerfaces. 2003;27:23–31. | |
Ladd J, Taylor AD, Piliarik M, Homola J, Jiang S. Label-free detection of cancer biomarker candidates using surface plasmon resonance imaging. Anal Bioanal Chem. 2009;393:1157–1163. | |
Chung J, Bernhardt R, Pyun J. Additive assay of cancer marker CA 19-9 by SPR biosensor. Sens Actuators B Chem. 2006;118:28–32. | |
Miao X, Guo X, Xiao Z, Ling L. Electrochemical molecular beacon biosensor for sequence-specific recognition of double-stranded DNA. Biosens Bioelectron. 2014;59:54–57. | |
Xuan F, Luo X, Hsing I-M. Ultrasensitive solution-phase electrochemical molecular beacon-based DNA detection with signal amplification by exonuclease III-assisted target recycling. Anal Chem. 2012;84:5216–5220. | |
Lin Y-H, Tseng W-L. A room-temperature adenosine-based molecular beacon for highly sensitive detection of nucleic acids. Chem Commun (Camb). 2012;48:6262–6264. | |
Tang Z, Liu P, Ma C, et al. Molecular beacon based bioassay for highly sensitive and selective detection of nicotinamide adenine dinucleotide and the activity of alanine aminotransferase. Anal Chem. 2011;83:2505–2510. | |
Xu H, Hepel M. “Molecular beacon”-based fluorescent assay for selective detection of glutathione and cysteine. Anal Chem. 2011;83:813–819. | |
Piao Y, Liu F, Seo TS. A novel molecular beacon bearing a graphite nanoparticle as a nanoquencher for in situ mRNA detection in cancer cells. ACS Appl Mater Interfaces. 2012;4:6785–6789. | |
Yu Y, et al. in American Physical Society March Meeting Abstracts. 2013;1:1029–1030. | |
Mathur A, Blais S, Goparaju CM, Neubert T, Pass H, Levon K. Development of a biosensor for detection of pleural mesothelioma cancer biomarker using surface imprinting. PLoS One. 2013;8:1–8. | |
Su L, Zou L, Fong CC, et al. Detection of cancer biomarkers by piezoelectric biosensor using PZT ceramic resonator as the transducer. Biosens Bioelectron. 2013;46:155–161. | |
Jain KK. Applications of nanobiotechnology in clinical diagnostics. Clin Chem. 2007;53:2002–2009. | |
Choi JS, Jun YW, Yeon SI, Kim HC, Shin JS, Cheon J. Biocompatible heterostructured nanoparticles for multimodal biological detection. J Am Chem Soc. 2006;128:15982–15983. | |
Xing Y, Chaudry Q, Shen C, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–1165. | |
Zhang Y, Wang T-H. Quantum dot enabled molecular sensing and diagnostics. Theranostics. 2012;2:631–654. | |
El-Sayed IH, Huang X, El-Sayed MA. Surface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: applications in oral cancer. Nano Lett. 2005;5:829–834. | |
Na HB, Lee JH, An K, et al. Development of a T1 contrast agent for magnetic resonance imaging using MnO nanoparticles. Angew Chem Int Ed Engl. 2007;119:5493–5401. | |
Ho YP, Kung MC, Yang S, Wang TH. Multiplexed hybridization detection with multicolor colocalization of quantum dot nanoprobes. Nano Lett. 2005;5:1693–1697. | |
Shanmukh S, Jones L, Driskell J, Zhao Y, Dluhy R, Tripp RA. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006;6(11):2630–2636. | |
Corsi F, De Palma C, Colombo M, et al. Towards ideal magnetofluorescent nanoparticles for bimodal detection of breast-cancer cells. Small. 2009;5:2555–2564. | |
Altintas Z, Uludag Y, Gurbuz Y, Tothill I. Development of surface chemistry for surface plasmon resonance based sensors for the detection of proteins and DNA molecules. Anal Chim Acta. 2012;712:138–144. | |
Medley CD, Smith JE, Tang Z, Wu Y, Bamrungsap S, Tan W. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal Chem. 2008;80:1067–1072. | |
Smith JE, Medley CD, Tang Z, Shangguan D, Lofton C, Tan W. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal Chem. 2007;79:3075–3082. | |
Farias PM, Santos BS, Fontes A. Semiconductor fluorescent quantum dots: efficient biolabels in cancer diagnostics. Methods Mol Biol. 2009;544:407–419. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.