Back to Journals » Cancer Management and Research » Volume 12
miR-665 Suppresses the Epithelial–Mesenchymal Transition and Progression of Gastric Cancer by Targeting CRIM1
Authors Wu KZ, Zhang CD, Zhang C, Pei JP, Dai DQ
Received 11 December 2019
Accepted for publication 21 April 2020
Published 15 May 2020 Volume 2020:12 Pages 3489—3501
DOI https://doi.org/10.2147/CMAR.S241795
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Kun-Zhe Wu,1 Chun-Dong Zhang,1,2 Cheng Zhang,1 Jun-Peng Pei,1 Dong-Qiu Dai1,3
1Department of Gastrointestinal Surgery, The Fourth Affiliated Hospital of China Medical University, Shenyang, Liaoning 110000, People’s Republic of China; 2Department of Gastrointestinal Surgery, Graduate School of Medicine, The University of Tokyo, Kashiwa 277-8561, Japan; 3Cancer Center, The Fourth Affiliated Hospital of China Medical University, Shenyang, Liaoning 110000, People’s Republic of China
Correspondence: Dong-Qiu Dai
Department of Gastrointestinal Surgery, The Fourth Affiliated Hospital of China Medical University, No. 4 Chongshan Road, Shenyang, Liaoning 110000, People’s Republic of China
Tel +86 159 0983 5960
Fax +86 24 6204 3110
Email [email protected]
Background: Gastric cancer (GC) is one of the most common aggressive cancers and is characterized by high mortality. Increasing evidence has shown that microRNA-665 (miRNA-665) serves as inhibiting-miRNA in cancers. However, the role of miR-665 in GC is yet unclear.
Methods: miR-665 was first analyzed using bioinformatics. Subsequent quantitative real-time PCR was used to detect miR-665 expression levels in different GC cell lines and tissues. The function of miR-665 in GC cells was determined via Cell Counting Kit 8, colony formation, wound healing, and transwell assays. Furthermore, Western blotting was utilized to measure the expression level of epithelial–mesenchymal transition (EMT)-related proteins. The target prediction and luciferase reporter assays were performed to confirm the binding between miR-665 and 3ʹ-UTR of the CRIM1 gene. In addition, rescue assays were used to determine whether CRIM1 upregulation abolished the inhibitory effect of miR-665.
Results: The expression of miR-665 was significantly decreased in GC patients and GC cell lines. Clinical and pathological analyses showed that the low expression of miR-665 was significantly associated with high TNM stage (P = 0.007), distant metastasis (P = 0.031), and poor differentiation (P = 0.029). Endogenic mimics of miR-665 remarkably suppressed GC cell proliferation, migration, invasion, and EMT in in vitro experiments. Inhibition of miR-665 expression induced the opposite effects. The results of the bioinformatics analysis and dual-luciferase assay showed that miR-665 targeted the 3ʹ-UTR of the CRIM1 gene. Rescue assays revealed that overexpression of CRIM1 attenuated the inhibitory effects of miR-665 in GC progression and EMT.
Conclusion: The overall study results demonstrated that miR-665 inhibits tumor progression and EMT in GC by targeting CRIM1, indicating that miR-665 might be a potential therapeutic target in the treatment of GC patients.
Keywords: gastric cancer, prognosis, miR-665, CRIM1, epithelial–mesenchymal transition
Introduction
Gastric cancer (GC) is one of the most aggressive digestive malignant tumors and the fifth most common cause of cancer-related death in the world.1 In 2018, 1,000,000 new diagnoses of GC and 783,000 deaths were estimated worldwide.2 Although various therapeutic strategies have been developed for GC, the prognosis for patients with advanced GC remains poor and the treatments are often ineffective.3–5 Thus, it is necessary to explore the potential biological molecular mechanisms of GC development and discover important biomarkers for early diagnosis and anti-cancer targeted therapies.
MicroRNAs (miRNAs) are key components of the conservative noncoding RNA family. Approximately 18–24 nucleotides in length, they regulate target mRNA expression via complementary pairing with the 3′-untranslated regions (3′-UTRs) of target mRNAs to mediate target gene degradation and regulate translation.6,7 Several studies have shown that miRNAs are frequently dysregulated in cancers, including GC. For example, miR-214 inhibited the tumor-promoting effect of cancer-associated fibroblasts (CAFs) on GC by targeting FGF9 in CAFs and also by regulating the EMT process in GC cells.8 Moreover, Li et al9 found that miR-188-5p was overexpressed in GC patients and promoted GC cell migration and invasion abilities by activating Wnt/β-catenin signaling. Furthermore, studies have shown that miRNAs regulate the biological behaviors of cancers, including proliferation, invasion, migration, and epithelial–mesenchymal transition (EMT).10–13 EMT serves as a key process in the pathogenesis of GC. In recent years, EMT has been reported to be regulated by miRNAs, such as miR-223,10 miR-630,14 and miR-345,15 suggesting that EMT-related miRNAs might serve as potential biomarkers and targets for therapies in distant metastasis of GC.
miR-665 is an intergenic miRNA, with its gene located on human chromosome 14q32.2. Recent research has revealed that miR-665 can regulate EMT-related proteins by targeting mRNAs in various cancers.16,17 It has also been reported that miR-665 is down-regulated in osteosarcoma,18 retinoblastoma,19 and ovarian cancer,20 but is up-regulated in non-small cell lung cancer21 and breast cancer.22 However, the functional role of miR-665 in GC remains unclear.
In this study, miR-665 expression and its prognostic value were first evaluated using bioinformatics analysis. Subsequently, miR-665 was demonstrated to be downregulated in GC tissues and cell lines, and associated with high TNM stage, distant metastasis, and poor differentiation. Functional experiments and Western blotting were performed to explore the miR-665 anti-cancer functions in GC. Then, the ability of miR-665 to inhibit migration, invasion, and EMT processes by directly targeting cysteine-rich motor neuron 1 (CRIM1) was verified through a series of experiments. Thus, it was revealed that miR-665 inhibits the EMT process and progression of GC by directly targeting CRIM1.
Materials and Methods
Bioinformatics Prediction
The gene expression profile microarray, GSE93415, was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and contained 20 pairs of GC tissues and adjacent normal tissues. Data were processed using the R software limma package to determine the different miRNA expression profiles. The cut-off criteria were P < 0.05 and |log2FC| > 1.0. Furthermore, RNA-seq and clinical GC data were downloaded from the Cancer Genome Atlas (TCGA) database to investigate the relationship between the expression level of miR-665 and GC patient survival. A total of 375 GC tissues and 32 normal gastric tissues were included in the study.
GC Cell Lines and Tissue Samples
Four human GC cell lines, including AGS, HGC-27, MKN-45, and MGC-803, and a normal gastric epithelial cell line (GES-1) were purchased from the Chinese Academy of Sciences (Shanghai, China). All cells were cultivated in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Invitrogen) and were maintained in a humidified atmosphere of 5% CO2 at 37°C. Sixty-three paired surgically-resected GC tissue and adjacent normal tissue (>5 cm from cancer tissue) samples were collected from the Fourth Affiliated Hospital of China Medical University, between November 2016 and June 2017. All tissues were snap-frozen in liquid nitrogen and promptly stored at –80°C after surgical removal. None of the patients enrolled in this study received preoperative chemotherapy and/or radiotherapy. Informed consent was obtained from all GC patients. TNM stage histological grade was confirmed based on the 8th American Joint Committee on Cancer (AJCC) system. The study was approved by The Medical Association Ethics Committee of the Fourth Affiliated Hospital of China Medical University.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
RNAios Plus (Takara Bio Inc., Shiga, Japan) was used to extract total RNA from cell lines and tissues, according to the manufacturer’s instructions. Reverse transcription and qRT-PCR of miR-665 were performed using the Hairpin-it™ miRNA RT-PCR Quantitation Kit (Gene Pharma, Shanghai, China), with U6 RNA as the internal reference. RNA reverse transcription was synthesized with the PrimerScriptTM reagent kit (Takara, Dalian, China) and SYBR Green (Solarbio, Beijing, China) was utilized to analyze the CRIM1 mRNA expression level, where glyceraldehyde phosphate dehydrogenase (GAPDH) was used as an endogenous control. The Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, US) was used to perform qRT-PCR. All primers were as follows: miR-665 sense, 5′-GGTGAACCAGGAGGCTGAGG-3′, miR-665 antisense, 5′-CAGTGCAGGGTCCGAGGTAT-3′, U6 sense, 5′-CGCTTCGGCAGCACATATAC-3′, U6 antisense, 5′-TTCACGAATTTGCGTGTCATC-3′, CRIM1 sense, 5′- AGTTTCCAAGTCAGGATATGTGC-3′, CRIM1 antisense, 5′- AGCATAACCCTCGATCAGAACA-3′, GAPDH sense, 5′-AGCCACATCGCTCAGACTC-3′, GAPDH antisense, 5′- GCCCAATACGACCAAATTC −3′.
Cell Transfection
The miR-665 mimics, mimic controls, miR-665 inhibitors, and inhibitor controls were synthesized by the GenePharma Company (Shanghai, China). In order to overexpress CRIM1, the CRIM1 coding sequence was inserted into the pcDNA3.1 eukaryotic expression plasmid (Invitrogen). Then, miR-665 mimics, mimic controls, miR-665 inhibitors, inhibitor controls, and CRIM1 and pcDNA3.1 plasmid were transfected using the Lipofectamine 3000 reagent (Invitrogen) into HGC-27 and MGC-803 cells, according to the manufacturer’s protocol.
Cell Proliferation Assays
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8) and colony formation assays. After a 24-h transfection with miRNA, 5×103 transfected cells were seeded into each well in 96-well plates with 100 μL of medium. After 0, 24, 48, 72, and 96 h of incubation, 10 μL of the CCK-8 solution (Solarbio) was added to each well and incubated at 37°C for 1 h. Results were detected by a microplate reader with absorbance at 450 nm. For the plate colony assays, approximately 2×103 transfected cells were inoculated into each well in six-well plates. After two weeks of culture, the cells were fixed with 4% paraformaldehyde, stained using 0.1% crystal violet (Solarbio) for 10–30 min, and photographed after rinsing. Each experiment was performed three times.
Wound Healing Assay
The transfected cells were cultured in six-well plates for 24 h. Then, a linear scratch was drawn with a 100 μL pipette tip. The cells were cultured at 37°C with 5% CO2 and observed and photographed with an inverted microscope (Leica, Wetzlar, HE, GER) at 0 and 48 h. Wound healing percentage was assessed using ImageJ software (US National Institutes of Health, Bethesda, MD, USA).
Transwell Assays
Approximately 2×104 transfected cells were plated into the upper insert of a transwell chamber (BD, Biosciences, San Jose, CA, USA), with or without Matrigel (BD), to examine their invasion and migration functions, respectively. The upper transwell chamber was treated with 200 μL of serum-free medium, while the lower chamber was filled with medium containing 20% FBS (Invitrogen) as a stimulus. After 24 h of culture at 37°C, cells were fixed and stained as described above. Cells that migrated to the bottom of the filter were photographed in randomly selected fields of view with an inverted microscope (Leica).
Target Gene Prediction for miR-665
Potential target genes for miR-665 were identified using three bioinformatics tools, including miRDB (http://www.mirdb.org/), TargetScan (http://www.targetscan.org/), and miRWalk (http://zmf.umm.uniheidelberg.de/apps/zmf/mirwalk2/). Subsequently, the GEPIA (Gene Expression Profiling Interactive Analysis) dataset (http://gepia.cancer-pku.cn/), which is a comprehensive website based on TCGA and GTEx databases, was used to predict differential expression of the target genes in GC and adjacent normal tissues.23
Luciferase Reporter Assays
Wild-type (wt) and mutant (mut) CRIM1 3′-UTRs were inserted into the pmiR-RB-REPORT (RiboBio, Guangzhou, China). Cells were seeded in 24-well plates and transfected with CRIM1-3′ UTR wt or mut vectors, and miR-665 mimic or miR-NC using Lipofectamine 3000 (Invitrogen). Luciferase activity was measured with a dual-luciferase assay kit (Promega, Madison, WI, USA), in accordance with the manufacturer’s instructions. Each assay was repeated three times.
Western Blotting
RIPA buffer (Beyotime, Shanghai, China) containing protease inhibitors was utilized to extract the total protein. Protein sample concentration was quantified with the BCA Protein Assay Kit (Takara, Shanghai, China). The extracted proteins were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electro-transferred to a polyvinylidene fluoride membrane (Takara). After blocking with 5% bovine serum albumin for 2 h at room temperature, the membranes were incubated at 4°C with primary antibodies diluted with primary antibody dilution buffer (Solarbio) as follows: CRIM1 (1:500 dilution; Abcam, Hong-Kong, China), E-cadherin (1:100 dilution; Abcam), vimentin (1:1500 dilution; Abcam), N-cadherin (1:1500 dilution; Abcam), and β-actin (1:5000 dilution; Abcam). After incubation with peroxidase-conjugated secondary antibodies (goat anti-rabbit, 1:5000 dilution; Proteintech, Chicago, IL, USA) at room temperature, the signal was detected using ECL chemiluminescent reagents (Pierce, Rockford, IL, USA). The abundance of target protein bands was measured and analyzed using the chemiluminescence Western blotting detection system (Tanon, Shanghai, China).
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 (SPSS Inc., Chicago, IL, USA) and Prism 7.0 (GraphPad, Inc., La Jolla, CA, USA) software. The chi-squared test or paired Student’s t-test were used to perform comparisons between two groups. Survival analysis was performed using the Kaplan–Meier method and compared using the Log rank test. All data are expressed as the mean ± standard deviation (SD). P-values < 0.05 or < 0.01 were considered statistically significant. All experiments were performed at least three times.
Results
miR-665 Is Downregulated in GC
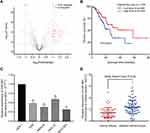
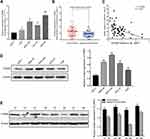
The GSE93415 dataset was downloaded from the NCBI-GEO database and analyzed using R software. A total of 110 abnormally expressed miRNAs were discovered (Supplementary Table 1). The 19 low-expression and 91 high-expression miRNAs are represented in a volcano plot (Figure 1A). MiR-665 was selected for further research after a comprehensive evaluation of the logFC value for each miRNA and of the reported studies. Subsequently, 375 samples from the TCGA were used to assess the association between miR-665 expression and GC patient survival. The Kaplan–Meier survival analysis indicated that a higher miR-665 expression is correlated with a higher survival rate in GC patients (P = 0.0249, Figure 1B).
Endogenous expression of miR-665 in four GC cell lines (AGS, HGC-27, MKN-45, and MGC-803) and in a normal gastric cell line (GES-1) was measured to further evaluate miR-665 expression. The qRT-PCR results indicated that miR-665 was significantly downregulated in four GC cell lines compared to GES-1, HGC-27 was relatively overexpressed, and MGC-803 was relatively downregulated (Figure 1C). In addition, miR-665 expression levels were detected in 63 GC tissues and corresponding adjacent normal tissues, revealing that miR-665 expression was diminished in GC tissues in comparison to the adjacent normal gastric tissues (Figure 1D). Clinical and pathological analyses showed that the low expression of miR-665 was significantly associated with high TNM stage (P = 0.007), distant metastasis (P = 0.031), and poor differentiation (P = 0.029, Table 1).
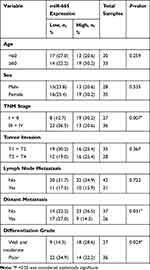 |
Table 1 Association Between miR-665 Expression and GC Clinicopathological Characteristics |
miR-665 Inhibits Proliferation of GC Cells
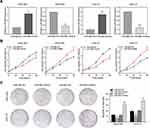
To elucidate the function of miR-665 in GC cells, relatively overexpressed HGC-27 and relatively downregulated MGC-803 were selected for the miR-665 transfection. Transfection efficiency was detected using qRT-PCR (Figure 2A). Subsequently, GC proliferation was examined using the CCK-8 assay. The results showed that the growth rate of HGC-27 and MGC-803 cells after miR-665 mimic transfection was significantly decreased, but increased upon the addition of miR-665 inhibitors (Figure 2B). Furthermore, colony formation assays indicated that cell proliferation was inhibited by miR-665 overexpression. Low expression of miR-665 showed the opposite effects (Figure 2C). These results demonstrated that miR-665 efficiently suppressed GC cell proliferation.
miR-665 Inhibits GC Cell Migration, Invasion, and EMT in vitro
Wound scratch and transwell assays were performed 24 h after transfection in an attempt to explore the effect of miR-665 on migration and invasion in GC cells. These results indicated that miR-665 overexpression inhibited GC cell migration, compared to the control group. MiR-665 expression inhibition showed the opposite effect (Figure 3A). Transwell results demonstrated that GC cells co-transfected with miR-665 mimics suppressed cell migration and invasion, while cells transfected with miR-665 inhibitors showed the opposite results (Figure 3B). Based on previous studies on the role of miR-665 in a variety of cancers, this study hypothesized that miR-665 might serve as an EMT-related miRNA. Hence, Western blotting was performed to detect EMT markers in MGC-803 and HGC-27 cells. The expression of the epithelial marker E-cadherin was enhanced after miR-665 upregulation in GC cells. However, the levels of mesenchymal markers N-cadherin and vimentin were decreased. Inhibition of miR-665 expression restrained E-cadherin expression and promoted N-cadherin and vimentin expression (Figure 3C). These findings indicated that miR-665 inhibited GC cell EMT process.
CRIM1 Is a Direct Target of miR-665
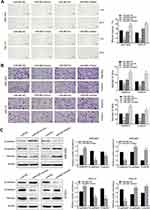
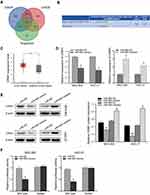
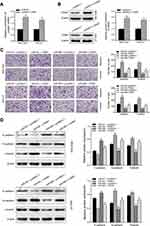
To explore the molecular mechanism of miR-665 in GC, three bioinformatics tools, including TargetScan7.1, miRDB, and miRWalk, were used to search for potential target genes of miR-665. A total of 117 target genes were selected (Figure 4A). Among the identified potential candidates, CRIM1 was chosen due to its high prediction score. Potential binding sites between CRIM1 and miR-665 were predicted using TargetScan7.1 (Figure 4B). Subsequently, GEPIA data were extracted to explore the expression of CRIM1. These results indicated that CRIM1 expression was significantly upregulated in GC tissues, compared to adjacent normal gastric tissues (P < 0.05, Figure 4C). Western blotting and qRT-PCR were performed to further verify the association between miR-665 and CRIM1. As a result, CRIM1 was downregulated and upregulated after transfection with miR-665 mimics and inhibitors, respectively (Figure 4D, E). The luciferase report assay showed that miR-665 mimics significantly inhibited the luciferase activity of the CRIM1-wt vector, but failed to significantly change the luciferase activity of the CRIM1-mut binding sites (Figure 4F).
CRIM1 expression level via qRT-PCR showed that CRIM1 was significantly upregulated in GC cell lines and tumor tissues (Figure 5A and B). Spearman correlation analysis indicated that CRIM1 expression was inversely correlated with miR-665 level in GC (Figure 5C). Moreover, Western blot results indicated that the CRIM1 protein level was upregulated in the GC cell lines and four tumor tissue samples (Figure 5D and E).
CRIM1 Attenuates miR-665 Inhibitory Effects on GC Cells
A rescue experiment was performed to confirm whether miR-665 suppresses GC cell EMT progress by regulating CRIM1. First, the pcDNA3.1-CRIM1 plasmid was transfected into MGC-803 and HGC-27 cells, and its transfection efficiency was examined by qRT-PCR and Western blot (Figure 6A and B). Subsequently, MGC-803 and HGC-27 cells were co-transfected with the pcDNA3.1-CRIM1 plasmid or empty vector and miR-665 mimics or negative controls to explore whether CRIM1 mediated the function of miR-665 in GC. Functional experimental results indicated that restoration of CRIM1 expression abrogated the inhibitory effects of miR-665 in relation to metastasis and invasion of GC cells (Figure 6C). Moreover, Western blot results revealed that co-transfection with CRIM1 partially abolished the regulatory effects of miR-665 on EMT marker expression in GC cells, including inhibition of E-cadherin and upregulation of N-cadherin and vimentin (Figure 6D). These results show that miR-665 inhibits GC progression and EMT through negative regulation of CRIM1.
Discussion
An increasing number of studies have reported that miRNAs play a crucial role in carcinogenesis and the progression of GC. However, comprehensive profiling and regulatory mechanisms of dysregulated miRNAs in cancer remain largely elusive. In the present study, the microarray database, GSE93415, was analyzed to discover that miR-665 was significantly down-regulated in GC cells. Clinical information and RNA-seq data from 375 GC patients downloaded from the TCGA demonstrated that GC patients with high miR-665 expression had a better prognosis than those with low expression. According to the qRT-PCR analysis, miR-665 was downregulated in GC cell lines and tissue specimens. In addition, clinicopathological features showed that downregulation of miR-665 correlated with a high TNM stage (P = 0.007), distant metastasis (P = 0.031), and poor differentiation (P = 0.029). Cell function experiments confirmed that miR-665 overexpression was able to inhibit GC cell proliferation, invasion, and migration in vitro, while low miR-665 expression showed the opposite effects.
Furthermore, Western blotting was performed to detect EMT-related markers in MGC-803 and HGC-27 cells transfected with miR-665 mimics or miR-665 inhibitor. Western blot results demonstrated that miR-665 participates in the process of EMT inhibition in GC. MiR-665 has been reported to significantly regulate the EMT process in various cancers. Liu et al previously concluded that miR-665 suppressed the development and metastasis of ovarian cancer by targeting homeobox A10.20 MiR-665 had a similar effect in colorectal cancer, inhibiting metastasis and EMT in vitro via the STAT3 signaling pathway.17 By contrast, miR-665 is up-regulated in hepatocellular carcinoma and miR-665 expression is associated with growth, metastasis, and EMT in hepatocellular carcinoma cells.24 Although miR-665 plays different roles in different cancers, these reports indicate that miR-665 can serve as an EMT-related miRNA. In recent years, EMT has been considered a prominent mechanism in the invasion and migration processes of tumors by restraining cell adhesion and facilitating cytoskeletal remodeling.25 However, the regulatory functions and mechanisms of EMT remain unclear, especially concerning miRNAs. The above results indicate that miR-665 is a key EMT-related miRNA in GC and requires further experimental research to explore its potential regulatory mechanisms.
The miRNAs have been previously confirmed to regulate target gene expression by binding to complementary sequences in the target gene 3′-UTR, leading to target gene degradation and translation inhibition. For example, miR-1224 functions as a tumor suppressor in intestinal GC by targeting focal adhesion kinase (FAK),26 while miR-1236-3p inhibits tumor metastasis in GC by targeting MTA2.27 In this study, bioinformatics tools and experimental assays were used to verify that the CRIM1 gene is the direct target of miR-665. CRIM1 is a glycosylated type I transmembrane protein that inhibits bone morphogenetic proteins (BMPs), maintains tissue architecture, and regulates cell migration and adhesion.28 A negative correlation between miR-665 and CRIM1 was also demonstrated in GC. CRIM1 has been reported to promote tumor metastasis and EMT in a variety of cancers and its expression is associated with tumor development and poor prognosis.29 Ogasawara et al found that CRIM1 activates invasion and EMT by downregulating E-cadherin and upregulating Claudin-1, matrix metalloproteinase (MMP) 2, and MMP9 in renal carcinoma cells.30 In addition, it has been reported that CRIM1 can promote EMT in lung and prostate cancers.31,32 In the present study, CRIM1 was shown to be upregulated in GC cell lines and tissues. Moreover, rescue assay results confirm that miR-665’s inhibitory effects are attenuated by CRIM1 in relation to progression and EMT of GC cells. These findings revealed that miR-665 inhibited the progression and EMT of GC by targeting CRIM1.
However, there are some limitations in the current study. First, in vivo experiments were not conducted to validate the findings. Second, this study did not involve investigating potential signaling pathways of miR-665 in regulatory mechanisms of GC. Furthermore, studies have shown that using histologically normal tissue adjacent to the tumor (NAT) as a healthy control tissue to characterize tumor-related biological processes and pathways may lead to suboptimal results.33,34 Since we collected and analyzed surgically-resected GC tissues and NAT, the prognostic value of miR-665 may not be adequately reflected. Hence, more experimental research is needed to further explore its diagnostic and therapeutic values.
In conclusion, this study demonstrated that miR-665 is downregulated in GC cell lines and tissues, and is associated with high TNM stage, distant metastasis, and poor differentiation. The results of a series of experiments indicated that miR-665 can inhibit GC cell proliferation, invasion, migration, and EMT by targeting CRIM1. The present study reveals that miR-665 and CRIM1 can serve as novel prognostic biomarkers of GC and are promising targets for GC therapy.
Ethics and Consent Statement
This study was approved by the Ethics Committee of the Fourth Affiliated Hospital, China Medical University in Liaoning Province, China. This study was conducted in accordance with the Declaration of Helsinki. We also obtained signed informed consents from all the patients.
Acknowledgments
This work was supported in part by Natural Science Foundation of Liaoning Province (No. 201602817), the Japan China Sasakawa Medical Fellowship (No. 2017816), and the China Scholarship Council (No. 201908050148).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548. doi:10.1001/jamaoncol.2016.5688
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Chau I, Fuchs CS, Ohtsu A, et al. Association of quality of life with disease characteristics and treatment outcomes in patients with advanced gastric cancer: exploratory analysis of RAINBOW and REGARD Phase III trials. Eur J Cancer. 2019;107:115–123. doi:10.1016/j.ejca.2018.11.013
4. Smyth EC, Cervantes A. Immunotherapy is not for all comers in chemotherapy-refractory advanced gastric cancer. Better predictive biomarkers are needed. Ann Oncol. 2018;29(10):2027–2028. doi:10.1093/annonc/mdy331
5. Li Y, Wei Y, He Q, Wang X, Fan C, Li G. Clinicopathological and prognostic significance of high circulating lymphocyte ratio in patients receiving neoadjuvant chemotherapy for advanced gastric cancer. Sci Rep. 2018;8(1):6223. doi:10.1038/s41598-018-24259-5
6. Shen L, Li Q, Wang J, et al. miR-144-3p promotes adipogenesis through releasing C/EBPalpha from Klf3 and CtBP2. Front Genet. 2018;9:677. doi:10.3389/fgene.2018.00677
7. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
8. Wang R, Sun Y, Yu W, et al. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res. 2019;38(1):20. doi:10.1186/s13046-018-0995-9
9. Li Y, Yan X, Shi J, et al. Aberrantly expressed miR-188-5p promotes gastric cancer metastasis by activating Wnt/beta-catenin signaling. BMC Cancer. 2019;19(1):505. doi:10.1186/s12885-019-5731-0
10. Hu J, Shan Z, Hu K, et al. miRNA-223 inhibits epithelial-mesenchymal transition in gastric carcinoma cells via Sp1. Int J Oncol. 2016;49(1):325–335. doi:10.3892/ijo.2016.3533
11. Zhang P, Tang WM, Zhang H, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117(4):525–534. doi:10.1038/bjc.2017.181
12. Yang D, Ma M, Zhou W, Yang B, Xiao C. Inhibition of miR-32 activity promoted EMT induced by PM2.5 exposure through the modulation of the Smad1-mediated signaling pathways in lung cancer cells. Chemosphere. 2017;184:289–298. doi:10.1016/j.chemosphere.2017.05.152
13. Wang M, Liu C, Su Y, et al. miRNA-34c inhibits myoblasts proliferation by targeting YY1. Cell Cycle. 2017;16(18):1661–1672. doi:10.1080/15384101.2017.1281479
14. Li D, Tian B, Jin X. miR-630 inhibits Epithelial-to-Mesenchymal Transition (EMT) by regulating the Wnt/beta-catenin pathway in gastric cancer cells. Oncol Res. 2018;27(1):9–17. doi:10.3727/096504018X15178732625479
15. Feng A, Yuan X, Li X. MicroRNA-345 inhibits metastasis and epithelial-mesenchymal transition of gastric cancer by targeting FOXQ1. Oncol Rep. 2017;38(5):2752–2760. doi:10.3892/or.2017.6001
16. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell Death Dis. 2018;9(6):706. doi:10.1038/s41419-018-0724-5
17. Ouyang S, Zhou X, Chen Z, Wang M, Zheng X, Xie M. LncRNA BCAR4, targeting to miR-665/STAT3 signaling, maintains cancer stem cells stemness and promotes tumorigenicity in colorectal cancer. Cancer Cell Int. 2019;19:72. doi:10.1186/s12935-019-0784-3
18. Dong C, Du Q, Wang Z, Wang Y, Wu S, Wang A. MicroRNA-665 suppressed the invasion and metastasis of osteosarcoma by directly inhibiting RAB23. Am J Transl Res. 2016;8(11):4975–4981.
19. Wang S, Du S, Lv Y, Zhang F, Wang W. MicroRNA-665 inhibits the oncogenicity of retinoblastoma by directly targeting high-mobility group box 1 and inactivating the Wnt/beta-catenin pathway. Cancer Manag Res. 2019;11:3111–3123. doi:10.2147/CMAR.S200566
20. Liu J, Jiang Y, Wan Y, Zhou S, Thapa S, Cheng W. MicroRNA665 suppresses the growth and migration of ovarian cancer cells by targeting HOXA10. Mol Med Rep. 2018;18(3):2661–2668. doi:10.3892/mmr.2018.9252
21. Li C, Qin F, Hu F, et al. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018;8:2. doi:10.1186/s13578-018-0202-x
22. Zhao XG, Hu JY, Tang J, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. 2019;10(7):479. doi:10.1038/s41419-019-1705-z
23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
24. Hu Y, Yang C, Yang S, Cheng F, Rao J, Wang X. miR-665 promotes hepatocellular carcinoma cell migration, invasion, and proliferation by decreasing Hippo signaling through targeting PTPRB. Cell Death Dis. 2018;9(10):954. doi:10.1038/s41419-018-0978-y
25. Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20(18):5403–5410. doi:10.3748/wjg.v20.i18.5403
26. Wang J, Wen T, Li Z, et al. MicroRNA-1224 inhibits tumor metastasis in intestinal-type gastric cancer by directly targeting FAK. Front Oncol. 2019;9:222. doi:10.3389/fonc.2019.00222
27. An JX, Ma MH, Zhang CD, Shao S, Zhou NM, Dai DQ. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018;18:66. doi:10.1186/s12935-018-0560-9
28. Wilkinson L, Kolle G, Wen D, Piper M, Scott J, Little M. CRIM1 regulates the rate of processing and delivery of bone morphogenetic proteins to the cell surface. J Biol Chem. 2003;278(36):34181–34188. doi:10.1074/jbc.M301247200
29. Zeng H, Tang L. CRIM1, the antagonist of BMPs, is a potential risk factor of cancer. Curr Cancer Drug Targets. 2014;14(7):652–658. doi:10.2174/1568009614666140725094125
30. Ogasawara N, Kudo T, Sato M, et al. Reduction of membrane protein CRIM1 decreases E-cadherin and increases Claudin-1 and MMPs, enhancing the migration and invasion of renal carcinoma cells. Biol Pharm Bull. 2018;41(4):604–611. doi:10.1248/bpb.b17-00990
31. Zeng H, Zhang Y, Yi Q, Wu Y, Wan R, Tang L. CRIM1, a newfound cancer-related player, regulates the adhesion and migration of lung cancer cells. Growth Factors. 2015;33(5–6):384–392. doi:10.3109/08977194.2015.1119132
32. Hudson BD, Hum NR, Thomas CB, et al. SOST inhibits prostate cancer invasion. PLoS One. 2015;10(11):e0142058. doi:10.1371/journal.pone.0142058
33. Russi S, Calice G, Ruggieri V, et al. Gastric normal adjacent mucosa versus healthy and cancer tissues: distinctive transcriptomic profiles and biological features. Cancers (Basel). 2019;11:9. doi:10.3390/cancers11091248
34. Aran D, Camarda R, Odegaard J, et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun. 2017;8(1):1077. doi:10.1038/s41467-017-01027-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.