Back to Journals » OncoTargets and Therapy » Volume 12
MiR-449a regulates the cell migration and invasion of human non-small cell lung carcinoma by targeting ADAM10
Authors Meng H, Huang Q, Zhang X, Huang J, Shen R, Zhang B
Received 9 October 2018
Accepted for publication 25 March 2019
Published 16 May 2019 Volume 2019:12 Pages 3829—3838
DOI https://doi.org/10.2147/OTT.S190282
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Haining Meng,1 Qiao Huang,1 Xijin Zhang,1 Jiawei Huang,1 Ruowu Shen,1 Bei Zhang2
1Department of Special Medicine, School of Basic Medical College, Qingdao University, Qingdao 266021, People’s Republic of China; 2Department of Immunology, School of Basic Medical College, Qingdao University, Qingdao 266021, People’s Republic of China
Background: MicroRNAs (miRNAs) are non-coding small RNAs that have been shown to play a key role in the development of many tumors. However, its specific mechanism of action in non-small cell lung cancer (NSCLC) is not very clear.
Purpose: This study was to identify the effect of miRNA-449a on NSCLC invasion and migration.
Methods: We used quantitative real-time PCR experiments to demonstrate that miRNA-449a is down-regulated in NSCLC tissues and cell lines. We also used the Transwell assay to detect cell invasion and migration, and the Western Blot assay was used to detect protein expression. The dual luciferase assay was used to detect the targeting relationship between miR-449a and A Disintegrin And Metalloproteinases 10 (ADAM10).
Results: Our experiments demonstrated that miRNA-449a was down-regulated in NSCLC tissues and cell lines. When miRNA-449a was up-regulated in NSCLC cells, the invasion and migration ability of the cells was weakened, and the expression of ADAM10 was decreased. After down-regulation of miRNA-449a, the cell’s invasion and migration ability was enhanced, and the expression of ADAM10 was increased. Through dual luciferase assays, we also found that miRNA-449a can target ADAM10 to delay the progression of epithelial-mesenchymal transition (EMT) and inhibit invasion and migration.
Conclusion: Our experiments demonstrated that miRNA-449a acted as a tumor suppressor gene through inhibiting the expression of ADAM10 in NSCLC.
Keywords: miR-449a, non-small cell lung cancer, NSCLC, ADAM10, epithelial-mesenchymal transition, EMT
Introduction
Lung cancer is one of the most common malignancies worldwide and is the leading cause of cancer deaths in both men and women.1,2 It is noteworthy that non-small cell lung carcinoma (NSCLC) accounts for about 80% of all lung cancer types.3,4 At present, the treatment of NSCLC with surgery has been greatly improved,5 however, the invasion and metastasis that has occurred at the time of diagnosis of NSCLC reduces the survival rate.6 There are many factors affecting the invasion and metastasis of NSCLC, but the signaling mechanism in tumor cells has not been clarified.
MicroRNAs (miRNAs) are endogenous non-coding RNA with small molecular weight and play an important role in animals and plants. It is one of the important gene regulatory molecules, which affects the gene expression of many proteins.7 Current research indicates that miRNAs affect tumor invasion, metastasis, proliferation, differentiation and apoptosis to some extent.8–10 Among them, miRNA-449a, a member of the miRNAs family, also plays an important role in the progress of the tumor. Studies have shown that, as a tumor suppressor, miRNA-449a promotes osteosarcoma apoptosis,11 inhibits proliferation of neuroblastoma cells,12 metastasis of liver cancer,13 and invasion of prostate cancer.14 So we wanted to explore the mechanism of how miRNA-449a affects NSCLC.
A disintegrin and metalloprotease (ADAM) are members of the metalloproteinase metzincin family and are approximately 750 amino acids in length.15,16 They affect intracellular protein interactions and post-translational modified expression.17 A Disintegrin And Metalloproteinases 10 (ADAM10) is an important member of the ADAM family of metalloproteinases and plays a key role in signaling inside and outside the cell.18,19 Cell adhesion is a must for many important functions of the human body. Cell adhesion disorder can lead to various pathological processes such as tumor invasion and metastasis. ADAM10 can regulate adhesion factors to affect the development of tumors, so ADAM10 plays an important role in tumor metastasis and invasion.20 There is an exciting increase of evidence that ADAM10 affects the metastasis and invasion of tumors by affecting epithelial-mesenchymal transition (EMT).21–23
The purpose of this experiment was to investigate the effect of miRNA-449a on the metastasis and invasion of NSCLC by regulating the ADAM10/EMT signaling pathway.
Materials and methods
Cell and tissues
Lung cancer cells (A549, H520, LH7) (Jikai Gene Company, Shanghai, People's Republic of China) were cultured in RPMI-1640 (HyClone, GE Healthcare Life Sciences, Marlborough, MA, USA), normal lung tissue cells (BEAS-2B) (Jikai Gene Company) were cultured in DMEM (HyClone) and 10% fetal bovine serum (ExCell Bio, Shanghai, People's Republic of China) was added. Trypsin-EDTA (Solarbio, Beijing, People's Republic of China) was used to digest parietal cells. The tissue samples were from Qingdao Municipal Hospital. The patient from the organization of the experiment signed a written consent form and the study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University. The tissue sample process was conducted in accordance with the Declaration of Helsinki.
Cell transfection
Overexpression of miRNA-449a (miRNA-449a mimic), down-regulation of miRNA-449a (miRNA-449a inhibitor) and corresponding control were synthesized by the Jikai Gene Company. A549 and H520 cells were seeded at 8,00,000 cells/hole in 6-well plates. After 24 hours, the medium in the 6-well plate was replaced with 1.5 ml of medium per well, and then the miRNA-449a mimic, miRNA-449a inhibitor, and control were mixed with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), respectively. They were then co-added to the corresponding 6-well plates and replaced with new medium after 6 hours. The cells were further cultured at 37°C in a 5% CO2 atmosphere. A549 was seeded in a 6-well plate at 800,000 cells/well. After 24 hours, the medium in the 6-well plate was replaced, and 1.5 ml of the medium was added to each well, after which the miRNA-449a inhibitor, control, miRNA-449a inhibitor + siRNA-ADAM10 (Dharmacon, Lafayette, CO, USA) were mixed with Lipofectamine 2000 (Thermo Fisher Scientific), respectively. Co-added to the corresponding 6-well plate and replaced with new medium after 6 hours. The cells were further cultured at 37°C in a 5% CO2 atmosphere.
miRNA-449a mimic:
ATTGTTAGCTGGCTGCTTGGGTCAAGTCAGCAGCCACAACTACCCTGCCACTTGCTTCTGGATAAATTCTTCTTGTCAATGAAGTGCTCTGGATACCTGTGTGTGATGAGCTGGCAGTGTATTGTTAGCTGGTTGAATATGTGAATGGCATCGGCTAACATGCAACTGCTGTCTTATTGCATATACAATGAACATCAGAGTGTAACTGAATCTGTAATTAGTGTGTGT
miRNA-449a inhibitor: ACCAGCTAACAATACACTGCCA
cDNA synthesis and quantitative real-time PCR
Total cellular RNA was extracted using the iso plus RNA extraction kit (Takara Bio Inc., Shiga, Japan). The reverse transcription reaction was carried out using PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Japan). Quantitative real-time PCR (qRT-PCR) (bio-rad CFX96) was used for SYBR Premix Ex Taq RR420A (Takara). β-actin is used for endogenous reference of target gene mRNA, and U6 small nuclear RNA is used for endogenous reference of miRNA. The primers used in the experiment were all purchased from Shanghai Shenggong.
miR-449a: Forward,TGGCAGTGTATTGTTA; Reverse, ATCCAGTGCAGGGTCCGAGG.
ADAM10: Forward, GGAGTGTACGTGTGCCAGTTCTG; Reverse, GGTTCGACCACTGAAGTGCCTAC.
E-cadherin: Forward, CTGATTCTGCTGCTCTTGCTG; Reverse, CTTCTCCGCCTCCTTCTTCA.
Vimentin: Forward, GAAATTGCAGGAGGAGATGC; Reverse, ATTCCACTTTGCGTTCAAGG.
U6: Forward, CTCGCTTCGGCAGCACA; Reverse, AACGCTTCACGAATTTGCGT.
β-actin: Forward, CATGTACGTTGCTATCCAGGC; Reverse, CTCCTTAATGTCACGCACGAT.
Transwell assays
Matrigel (Corning Incorporated, Corning, NY, USA) was mixed with complete medium at 1:9 and added to a Transwell (Corning) chamber and placed in a 37°C incubator for 4 hours. Only the Transwell chamber of the invasive test needed to have matrigel added. This step was not required for the migration test. A cell counting plate was used to count the number of cells. Then, the cells were seeded into the chamber at 50,000 cells/well and 200 μl of complete medium was added to the upper chamber, while 500 μl of 15% fetal bovine serum medium was added to the lower chamber. This was placed in a 37°C incubator for 24 hours, then taken out of the Transwell chamber and the inner membrane was wiped with a cotton swab. The medium was then fixed with methanol (Solarbio) for 10 mins, stained with 0.2% crystal violet (Solarbio) for 10 min, washed three times with PBS (Solarbio), and photographed with a microscope.
Dual luciferase reporter assay
The gene sequence of ADAM10 binding to miR-449a was predicted by software (TargetScan Human 7.1), and the gene sequence was inserted into the luciferase vector and named as ADAM10 3’UTR (Sauer Biotechnology Inc, People's Republic of China). miRNA-449amimic, miR-449ainhibitor, control (Jikai Gene Company) and fluorescent plasmid using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific)co-transfected into A549 cells. After 48 hours, the protein was extracted, and then the activity of luciferase was detected on a microplate reader using a dual luciferase kit (Promega Corporation, Fitchburg, WI, USA).
Western blot
Whole protein extraction was performed with RIPA lysis (Beyotime, Shanghai, People's Republic of China) buffer and protease inhibitors (Solarbio) cleavage on ice for 30 minutes. The lysate was centrifuged at 12,000 x g for 10 minutes, and the supernatant was the total protein. The extracted protein was mixed with a loading buffer SDS-PAGE Loading Buffer (CWBIO, Beijing, People's Republic of China) and boiled in water for 5 minutes. The sample was added to the prepared concentrated separation gel for electrophoresis. Afterwards, the protein bands in the gel were transferred to a PVDF membrane (Millipore, Billerica, MA, USA). At 4°C with the primary antibodies of ADAM10 (1:1000, Cell Signaling Technology (CST), Danvers, MA, USA), β-actin (1:1000, Bioss, Beijing, People's Republic of China), E-cadherin (1:1000, CST), Vimentin (1:1000, CST) were incubated overnight. And then incubated with secondary antibodies (1:3000, Bioss) at 25°C for 60 min at low speed of 70 rpm. Finally, imaging analysis was performed using the Quant LAS 4010 imaging system.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software. The results of the measurement data were expressed as mean ± standard deviation, and the data were compared using paired t test. The difference was significant at P<0.05.
Results
miR-449a expression is downregulated in NSCLC tissues and cell lines
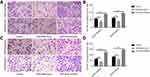
First, we used qRT-PCR to measure the expression of miRNA-449a in 69 pairs of NSCLC tissues and normal tissues. We found that the expression of miR-449a was only related to the disease stage (Table 1). Compared to the normal lung tissues, miRNA-449a is downregulated in NSCLC tissues (Figure 1A). Similarly, we used qRT-PCR to measure the expression of miRNA-449a in NSCLC cell lines (A549, H520, LH7) and normal lung tissue cell line BEAS-2B. Compared to the normal lung tissue cell line BEAS-2B, miRNA-449a is under-represented in three NSCLC cells (Figure 1B). From the above experimental results, it could be concluded that miRNA-449a had low expression in NSCLC tissues and cell lines.
 | Table 1 Clinicopathological Features |
Successfully altered miR-449a expression in A549 and H520 cell lines
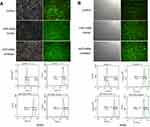
The constructed control, miR-449a mimic and miR-449a inhibitor were successfully transferred into A549 and H520 cell lines by the transfection technique. In A549 cells, the transfection efficiency of the control group, miR-449a mimic and miR-449a inhibitor was over 99% (Figure 2A). In H520 cells, the transfection efficiency of the control group, miR-449a mimic and miR-449a inhibitor was over 98% (Figure 2B).
The expression level of miR-449a in A549 cells was then examined by qRT-PCR. Compared with the control group, miR-449a increased expression in the miR-449a mimic group and decreased expression in the miR-449a inhibitor group (Figure 3A). Similarly, qRT-PCR was used to measure the expression level of miR-449a in H520 cells. Compared with the control group, miR-449a increased expression in the miR-449a mimic group and decreased expression in the miR-449a inhibitor group (Figure 3B). It was confirmed by testing that we successfully changed the expression level of miR-449a in A549 and H520 cell lines by transfection technology.
 | Figure 3 Successfully up-regulated and down-regulated miR-449a in A549 and H520 cell lines.Notes: (A) Changes of miR-449a in A549 cell lines. (B) Changes of miR-449a in H520 cell lines. **p<0.01. |
miR-449a inhibits the migration and invasion of NSCLC
In the A549 (Figure 4A and B) and H520 (Figure 4C and D) cell lines, compared with the control group, the invasion and migration ability of the cells in the miR-449a mimic group was weakened, and the invasion and migration ability of the cells in the miR-449a inhibitor group was enhanced. This demonstrates that miR-449a inhibits the invasion and migration of NSCLC.
miR-449a regulates EMT by targeting ADAM10
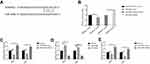
In order to prove that miR-449a affects the invasion and migration ability of NSCLC, we first used informatics analysis software (TargetScan Human 7.1) to search for direct target protein of miR-449a, and found that some gene sequences of ADAM10 may directly bind to miR-449a (Figure 5A). To confirm whether miR-449a is determined to bind to ADAM10, we performed a dual luciferase assay. In the dual luciferase assay, the ADAM103’UTR plasmid was co-transfected with miR-449a mimic control, miR-449a mimic, miR-449a inhibitor control, and miR-449a inhibitor. Compared with the control group, the fluorescence intensity of the miR-449a mimic group was weakened, and the fluorescence intensity of the miR-449a inhibitor group was enhanced (Figure 5B). The above results demonstrate that miR-449a is targeted to bind ADAM10. The expression levels of ADAM10, E-cadherin and Vimentin in control, miR-449a mimic and miR-449a inhibitor groups of A549 and H520 cell lines were measured by qRT-PCR. Compared with the control group, the expression level of ADAM10 (Figure 5C) and Vimentin (Figure 5E) was decreased, while the expression level of E-cadherin was increased (Figure 5D) in the miR-449a mimic group. Compared with the control group, the expression of ADAM10 (Figure 5C) and Vimentin (Figure 5E) was increased, while the expression of E-cadherin was decreased (Figure 5D) in the miR-449a inhibitor group.
Then we found that in the A549 (Figure 6A) and H520 (Figure 6B) cell lines, compared with the control group, the expression of ADAM10 was decreased, and the expression of EMT-related protein E-cadherin was increased, Vimentin was reduced in the miR-449a mimic group (Figure 6C and E). Compared with the control group, the expression of ADAM10 was increased, the expression of E-cadherin was decreased, and the expression of Vimentin was increased in the miR-449a inhibitor group (Figure 6D and F). From Figures 5 and 6, it can be concluded that miR-449a can regulate EMT by targeting ADAM10.
miR-449a regulates invasion and migration of NSCLC by targeting ADAM10
To further determine that miR-449a is capable of regulating the invasion and migration of NSCLC by ADAM10, we used siRNA-ADAM10 to silence the expression of ADAM10. The siRNA-ADAM10 and miR-449a inhibitor were co-transfected into A549 cells. Compared with the miR-449a inhibitor group, the cell invasion, migration ability (Figure 7A and B) and ADAM10, E-cadherin, and Vimentin protein expression levels (Figure 7C and D) occurred reply phenomenon in the siRNA-ADAM10 and miR-449a inhibitor co-transfection group. This indicates that mir-449a plays an important role in the invasion and migration of NSCLC by targeting ADAM10 to regulate EMT.
Discussion
MicroRNAs (miRNAs) are small molecular weight non-coding RNA,7 and there is increasing evidence that miRNAs play an important role in the development of tumors, and it is also a popular research direction of anticancer drug targets. miR-198 can inhibit the migration of diffuse large B-cell lymphoma by targeting HMGA1.24 miR-18a24 and miR-18b regulate ORAI3 by positive targeting, and miR-34a regulate ORAI3 by negative targeting in order to regulate tumor progression.25 miR-346 can inhibit the development of liver cancer by targeting inhibition of KLF14.26 In the miRNAs family, each miRNA plays its part, and the current research proves that miR-449a plays a role as a tumor suppressor gene in most tumors, inhibiting tumor invasion and metastasis and so on. For example, miR-499a can decelerate glioma cell proliferation while accelerating apoptosis by targeting inhibition of Notch1.27 Furthermore, miR-449a can inhibit the growth of liver cancer by targeting c-MET, and miR-449a can also inhibit the invasion and metastasis of NSCLC by targeting c-MET.28,29 Our study is consistent with these studies, miR-449a plays a role as a tumor suppressor gene. In the experiment, we found that the expression of miR-449a has no relationship with the patient’s gender, age, lymphonode migration and tumor histology subtype, but it is related to the disease stage. We demonstrated that miR-449a can bind to ADAM10 by dual luciferase assay, and it was confirmed that miR-449a negatively regulates ADAM10 to inhibit the invasion and migration ability of NSCLC by qRT-PCR, Transwell and Western Blot. Based on the existing evidence and our experimental results, miR-449a is an important target for slowing the invasion and migration of NSCLC.
ADAM10 is highly expressed in many different types of tumors and has important clinical significance. It can regulate cell migration, extracellular signal transduction, and cell adhesion.15,30 ADAM10 is highly expressed in cancer cells that can promote the metastasis of colorectal cancer,31 it can promote the carcinogenesis and cell proliferation of oral squamous cell carcinoma,32 and can promote the invasion of prostate cancer.33 Furthermore, the current study demonstrates that ADAM10 is highly expressed in NSCLC and can promote the development of NSCLC and can be used as a tumor biomarker for NSCLC.34,35 Therefore, it is meaningful to select ADAM10 as a target of miR-449a in NSCLC. The purpose of this study was to identify the effect of miR-449a on NSCLC invasion and migration, but EMT is recognized as an indispensable condition for tumor invasion and migration.36 Studies have shown that KMT2C can promote the progression of EMT and promote the occurrence of invasion and metastasis in gastric cancer,37 activation of the NFKB-SNAI1 signaling pathway can induce EMT of cancer cells and promote metastasis of liver cancer,38 TFF1 can slow the invasive transformation by preventing EMT of pancreatic intraepithelial neoplasia.39 The current evidence suggests that since we study the invasion and migration of NSCLC, it is necessary to study EMT. Moreover, previous studies have also proved that ADAM10 is involved in the development of EMT.40–43
In our experiments, after up-regulation of miR-449a, the expression of ADAM10 was decreased, the expression of EMT-related major protein E-cadherin was increased, and the expression of Vimentin was decreased. After down-regulating miR-449a, the opposite is true. It was shown that with the change of miR-449a in NSCLC, ADAM10, E-cadherin, Vimentin also changed, being negatively correlated with ADAM10, Vimentin, and positively correlated with E-cadherin. Combined with dual luciferase experiments, it was demonstrated that miR-449a can target ADAM10 to affect the invasion and migration of NSCLC. However, this does not prove that the effect of miR-449a on NSCLC invasion and migration is the result of ADAM10 regulating EMT. Next, our reply experiment gave the answer. The experimental results confirmed that ADAM10 can reverse the effect of miRNA-449a on the invasion and migration of NSCLC, and also reverse the expression of E-cadherin and Vimentin. This indicates that miR-449a does inhibit the invasion and migration of NSCLC by targeting ADAM10 to influence the progression of EMT. Liu et al's44 team has demonstrated that miR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma. It is indicated that miR-449a not only plays a role as a tumor suppressor gene in NSCLC, but also inhibits the invasion and proliferation of liver cancer. This provides additional evidence for the inhibition of tumor progression by miR-449a targeting ADAM10.
Finally, we have shown that miR-449a inhibits the invasion and migration of NSCLC by targeting ADAM10 to modulate EMT. MiR-449a is of great interest as a potential target for the study of NSCLC. However, the specific mechanisms of miR-449a on NSCLC still have many ambiguous aspects. It is believed that miR-449a can affect the invasion and migration ability of NSCLC by other pathways. This requires us to continue our studies in the future.
Acknowledgments
This work was supported by Shandong Provincial Natural Science Foundation of China (No.J16LK04).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jamal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19.
3. Ou SHI, Zell JA, Ziogas A, Anton‐Culver H. Prognostic factors for survival of stage I nonsmall cell lung cancer patients. Cancer. 2007;110(7):1532–1541. doi:10.1002/cncr.22812
4. Spira A, Ettinger DS. Multidisciplinary management of lung cancer. New Engl J Med. 2004;350(19):2008. doi:10.1056/NEJMra035536
5. De LP, Decker G. [Surgical treatment of non-small cell lung cancer]. Rev Mal Respir. 2004;21(5 Pt 1):971.
6. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
8. Li X, Zhang Y, Zhang H, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9(7):824–833. doi:10.1158/1541-7786.MCR-10-0529
9. Li N, Nan C, Zhong X, et al. miR-182-5p promotes growth in oral squamous cell carcinoma by inhibiting CAMK2N1. Cell Physiol Biochem. 2018;49(4):1329–1341. doi:10.1159/000493411
10. Zhou F, Yuan Q, Zhang W, et al. MiR-663a stimulates proliferation and suppresses early apoptosis of human spermatogonial stem cells by targeting NFIX and regulating cell cycle. Mol Ther Nucleic Acids. 2018;12:319–336. doi:10.1016/j.omtn.2018.05.015
11. Chen J, Zhou J, Chen X, et al. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol. 2015;36(10):8221–8229. doi:10.1007/s13277-015-3568-y
12. Xiao-Yan XU, Xiao-Shan LI. Over-expressed miR-449a inhibits proliferation and induces apoptosis of SH-SY5Y cells in vitro. Basic Clin Med. 2017;8:1117–1121.
13. Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. doi:10.1016/j.jhep.2017.01.004
14. Kumar P, Sharad S, Petrovics G, et al. Loss of miR-449a in ERG-associated prostate cancer promotes the invasive phenotype by inducing SIRT1. Oncotarget. 2016;7(16):22791–22806. doi:10.18632/oncotarget.8061
15. Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi:10.1101/gad.1039703
16. Huovila A, Turner AH. M, Karkkainen L, Ortiz R. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30(7):413–422. doi:10.1016/j.tibs.2005.05.006
17. Tousseyn T, Jorissen E, Reiss K, Hartmann D. (Make) stick and cut loose–disintegrin metalloproteases in development and disease. Birth Defects Res C Embryo Today. 2010;78(1):24–46. doi:10.1002/bdrc.20066
18. Reiss K, Maretzky T, Ludwig A, et al. ADAM10 cleavage of N-cadherin and regulation of cell|[ndash]|cell adhesion and |[beta]|-catenin nuclear signalling. Embo J. 2014;24(4):742–752. doi:10.1038/sj.emboj.7600548
19. Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139(20):3693–3709. doi:10.1242/dev.076398
20. Reiss K, Ludwig A, Saftig P. Breaking up the tie: disintegrin-like metalloproteinases as regulators of cell migration in inflammation and invasion. Pharmacol Ther. 2006;111(3):985–1006. doi:10.1016/j.pharmthera.2006.02.009
21. Hou L, Du Y, Zhao C, Wu Y. PAX2 may induce ADAM10 expression in renal tubular epithelial cells and contribute to epithelial-to-mesenchymal transition. Int Urol Nephrol. 2018;50(9):1729–1741. doi:10.1007/s11255-018-1956-0
22. Park G, Kim D. TLR4-mediated galectin-1 production triggers epithelial-mesenchymal transition in colon cancer cells through ADAM10- and ADAM17-associated lactate production. Mol Cell Biochem. 2017;425(1–2):191–202. doi:10.1007/s11010-016-2873-0
23. Guo J, He L, Yuan P, et al. ADAM10 overexpression in human non-small cell lung cancer correlates with cell migration and invasion through the activation of the Notch1 signaling pathway. Oncol Rep. 2012;28(5):1709–1718. doi:10.3892/or.2012.2003
24. Wang R, Shen J, Wang Q, Zhang M. Bortezomib inhibited the progression of diffuse large B-cell lymphoma via targeting miR-198. Biomed Pharmacother. 2018;108:43–49. doi:10.1016/j.biopha.2018.08.151
25. Vashisht A, Tanwar J, Motiani R. Regulation of proto-oncogene Orai3 by miR18a/b and miR34a. Cell Calcium. 2018;75:101–111. doi:10.1016/j.ceca.2018.08.006
26. Wang Y, Liu J, Shi M, Chen F. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J Cell Physiol. 2018;234(1):572–580.
27. Wang B, Yang B, Yang H, et al. MicroRNA-499a decelerates glioma cell proliferation while accelerating apoptosis through the suppression of Notch1 and the MAPK signaling pathway. Brain Res Bull. 2018;142:96–106. doi:10.1016/j.brainresbull.2018.06.005
28. Cheng J, Wu L, Deng X, et al. MicroRNA-449a suppresses hepatocellular carcinoma cell growth via G1 phase arrest and the HGF/MET c-Met pathway. Hepatobiliary Pancreat Dis Int. 2018;17(4):336–344. doi:10.1016/j.hbpd.2018.07.006
29. Luo W, Huang B, Li Z, et al. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One. 2013;8(5):e64759. doi:10.1371/journal.pone.0064759
30. Wolfsberg T, Primakoff P, Myles D, White J. ADAM, a novel family of membrane proteins containing A disintegrin and metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131(2):275–278.
31. Gavert N, Sheffer M, Raveh S, et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67(16):7703–7712. doi:10.1158/0008-5472.CAN-07-0991
32. Ko S, Lin S, Wong Y, Liu C, Chang K, Liu T. Increase of disintergin metalloprotease 10 (ADAM10) expression in oral squamous cell carcinoma. Cancer Lett. 2007;245(1–2):33–43. doi:10.1016/j.canlet.2005.10.019
33. McCulloch D, Akl P, Samaratunga H, Herington A, Odorico D. Expression of the disintegrin metalloprotease, ADAM-10, in prostate cancer and its regulation by dihydrotestosterone, insulin-like growth factor I, and epidermal growth factor in the prostate cancer cell model LNCaP. Clin Cancer Res. 2004;10(1 Pt 1):314–323.
34. Atapattu L, Saha N, Chheang C, et al. An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth. J Exp Med. 2016;213(9):1741–1757. doi:10.1084/jem.20151095
35. Yoneyama T, Gorry M, Sobo-Vujanovic A, et al. ADAM10 sheddase activity is a potential lung-cancer biomarker. J Cancer. 2018;9(14):2559–2570. doi:10.7150/jca.24601
36. Campbell K. Contribution of epithelial-mesenchymal transitions to organogenesis and cancer metastasis. Curr Opin Cell Biol. 2018;55:30–35. doi:10.1016/j.ceb.2018.06.008
37. Cho S, Yoon C, Lee J, et al. KMT2C mutations in diffuse type gastric adenocarcinoma promote epithelial-to-mesenchymal transition. Clin Cancer Res. 2018. doi:10.1158/1078-0432.CCR-17-1679
38. Chen D, Ning W, Li X, et al. Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma. Autophagy. 2018;14(8):1335–1346. doi:10.1080/15548627.2018.1474994
39. Yamaguchi J, Yokoyama Y, Kokuryo T, Ebata T, Enomoto A, Nagino M. Trefoil factor 1 inhibits epithelial-mesenchymal transition of pancreatic intraepithelial neoplasm. J Clin Invest. 2018;128(8):3619–3629. doi:10.1172/JCI97755
40. Schirrmeister W, Gnad T, Wex T, et al. Ectodomain shedding of E-cadherin and c-Met is induced by Helicobacter pylori infection. Exp Cell Res. 2009;315(20):3500–3508. doi:10.1016/j.yexcr.2009.07.029
41. Doberstein K, Pfeilschifter J, Gutwein P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011;32(11):1713–1723. doi:10.1093/carcin/bgr195
42. Park G, Kim D, Kim Y, et al. Regulation of ADAM10 and ADAM17 by Sorafenib inhibits epithelial-to-mesenchymal transition in epstein-barr virus-infected retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2015;56(9):5162–5173. doi:10.1167/iovs.14-16058
43. Park G, Kim D. Insulin-like growth factor-1 activates different catalytic subunits p110 of PI3K in a cell-type-dependent manner to induce lipogenesis-dependent epithelial-mesenchymal transition through the regulation of ADAM10 and ADAM17. Mol Cell Biochem. 2018;439(1–2):199–211. doi:10.1007/s11010-017-3148-0
44. Liu S, Liu K, Zhang W, et al. miR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res. 2016;8(6):2609–2619.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.