Back to Journals » Cancer Management and Research » Volume 12
miR-331-3p Inhibits Tumor Cell Proliferation, Metastasis, Invasion by Targeting MLLT10 in Non-Small Cell Lung Cancer
Authors Tian QQ, Xia J, Zhang X , Gao BQ, Wang W
Received 13 February 2020
Accepted for publication 24 June 2020
Published 14 July 2020 Volume 2020:12 Pages 5749—5758
DOI https://doi.org/10.2147/CMAR.S249686
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Qing-Qing Tian,1 Jing Xia,2 Xin Zhang,3 Bao-Qin Gao,4 Wei Wang5
1Department of Pathology, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 2General Department of Houhu, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 3Department of Radiology, The Fourth People’s Hospital of Huai’an, Huai’an, Huai’an, People’s Republic of China; 4Operating Room, Huai’an Second People’s Hospital and the Affiliated Huai’an Hospital of Xuzhou Medical University, Huai’an, People’s Republic of China; 5Department of Oncology, Huai’an Second People’s Hospital and the Affiliated Huai’an Hospital of Xuzhou Medical University, Huai’an, People’s Republic of China
Correspondence: Bao-Qin Gao
Operating Room, Huai’an Second People’s Hospital and the Affiliated Huai’an Hospital of Xuzhou Medical University, Huai’an, People’s Republic of China
Email [email protected]
Wei Wang
Department of Oncology, Huai’an Second People’s Hospital and the Affiliated Huai’an Hospital of Xuzhou Medical University, Huai’an, People’s Republic of China
Email [email protected]
Objective: Mounting research has established the role of microRNAs (miRNAs) as oncogenes or anti-oncogenes (tumor suppressors) in the development and progression of several cancers. The purpose of our current study is to delineate the roles and functional mechanisms of miR-331-3p and MLLT10 in non-small cell lung cancer (NSCLC) tumorigenesis.
Patients and Methods: Quantitative reverse transcription-polymerase chain reaction (RT-qPCR) was employed to measure miR-331-3p expression levels in twenty-six matched tumor tissues and non-cancerous tissues collected from patients suffering from NSCLC, and from six NSCLC cell lines separately: A549, H1650, H292, H1299, H1944 and BEAS-2b. We employed the dual-luciferase activity assay to check whether the putative gene, MLLT10, was a downstream target of miR-331-3p in NSCLC pathogenesis and development. Western blot was conducted to analyze the protein expression levels of MLLT10 (AF10), E-cadherin, Vimentin, and GAPDH. CCK-8 assay, transwell migration assay, and transwell invasion assay were carried out to observe the functions of miR-331-3p and MLLT10 on NSCLC tumor cell proliferation, metastasis, and invasion, respectively. To identify whether the metastasis of NSCLC tumor cells was EMT-mediated, supplementary experiments involving E-cadherin and Vimentin were implemented.
Results: miR-331-3p was downregulated in NSCLC, which promoted tumor cell proliferation, whereas the overexpression of miR-331-3p inhibited tumor cell proliferation. Being a direct target of miR-331-3p, MLLT10 was negatively modulated by miR-331-3p, which suppressed tumor cell proliferation, migration, and invasion in NSCLC. However, MLLT10 overexpression alleviated the above inhibitory effects. Furthermore, EMT-mediated metastasis was proved to be present in NSCLC.
Conclusion: miR-331-3p played a suppressor role in NSCLC tumor cell proliferation, EMT-mediated metastasis, and invasion by targeting MLLT10. Our findings highlighted that miR-331-3p/MLLT10 axis could be useful as a clinical diagnostic marker and therapeutic target in NSCLC patients.
Keywords: NSCLC, miR-331-3p, MLLT10, EMT-mediated metastasis
Introduction
Non-small cell lung cancer (NSCLC) accounts for about four-fifths of lung cancers,1 and is the most lethal of all carcinomas in the world, with 11.6% morbidity rate and 18.4% mortality rate, according to GLOBOCAN 2018. Smoking is considered to be the most common risk factor for NSCLC, followed by genetic variations, which is also responsible for lung cancer.2,3 Due to the high recurrence rate and greater migrate, the 5-year survival rate is maintained at a lower level, ie, about 10–20%, despite great improvements in the detection and treatment of lung cancer in the last few decades.4,5 In recent years, a large number of studies have been conducted to find more potential biomarkers, like genes and proteins, which are involved in the development of NSCLC. Moreover, several microRNAs have been reported to be associated with NSCLC.
MicroRNAs (miRNAs) are a group of small, endogenous, non-coding RNA molecules, about 22 nucleotides in length.6 miRNAs exert regulatory effects on gene expression by binding to the 3ʹUTR region of the target mRNA transcribed by protein-coding genes, thus degrading the mRNA and preventing protein expression.7 Emerging evidence has illustrated that aberrations in the expression of miRNAs have been observed in several pathological diseases including malignant carcinomas, which produce a marked effect on the development of tissues and organs, especially in various cellular processes, including cell proliferation, metastasis, apoptosis, differentiation and metabolism.8,9 For example, Vannini et al summarized 37 miRNAs that were dysregulated in lung cancer, of which 24 miRNAs were associated with NSCLC.10 In addition, miR-31 was verified to play the role of an oncogene in NSCLC.11 In contrast, microRNA-29c, microRNA-30a, microRNA-216b, microRNA-944, microRNA-101, microRNA-154 and microRNA-185 functioned as anti-oncogenes in NSCLC.12–18 However, the functional role of miR-331-3p in NSCLC is not very clear and needs further investigation.
The MLLT10 gene, located at 10p13, encodes AF10 protein, which is known as a transcriptional activator of gene expression.19 Based on previous studies, MLLT10 gene has been reported as a crucial fusion partner gene in specific leukemic fusions; one of its frequent fusion genes is mixed-lineage leukemia (MLL, also known as KMT2A) detected in acute leukemias, with the translocation t(10;11)(p13;q23).20,21 Another common fusion gene is phosphatidylinositol-binding clathrin assembly lymphoid–myeloid (PICALM, previously called as CALM), with the translocation t(10;11)(p13;q21).22 In addition, NAP1L1-MLLT10 recombination was found in pediatric T-acute lymphoblastic leukemia.23 MLLT10/IL3 rearrangement was also found in acute lymphoblastic leukemia.24 Whether MLLT10 also plays a role in pathological diseases other than leukemias had piqued our interest.
The purpose of our study was to delineate the functional roles of miR-331-3p and MLLT10 in NSCLC tumorigenesis, which may provide useful information for NSCLC clinical diagnosis and treatment.
Patients and Methods
Samples and Tissues
Twenty-six cases of NSCLC patients who were admitted to our hospital from October 2017 to January 2019 were selected to conduct our study. Tumor tissues and paired non-cancerous tissues were collected from the 26 patients, who had not undergone any therapy. Total tissues were snap-frozen in liquid nitrogen and stored at –80 °C for later experiments. The clinical characteristics of the patients are summarized in Table 1. All the procedures followed for extraction and utilization of tissue samples complied with the ethical standards of our hospital. This study was approved by the ethics committee of Huai’an Second People’s Hospital and The Affiliated Huai’an Hospital of Xuzhou Medical University. All patients provided their informed consent before participating in the study.
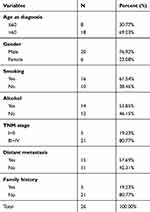 |
Table 1 Clinical Characteristics of the Study Participants |
Cell Culture
Six types of HSCLC cell lines: BEAS-2b, H1299, A549, H1650, H292and H1944 were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Roswell Park Memorial Institute (RPMI-1640) medium, supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS), in humidified incubators with 5% CO2 at 37 °C.
Transfection
miR-331-3p inhibitor (inhibitor-miR-331-3p) or its negative control (inhibitor-NC) was transfected into A549 and H1944 cells separately, whereas miR-331-3p mimic (mimic-miR-331-3p) or its negative control (mimic-NC) was transfected into H1650 and H1299 cells separately to observe the functions of miR-331-3p on tumor cell proliferation. Luciferase activity was measured after the transfection of mimic-miR-331-3p and mimic-NC into H1650 and H1299 cells separately. LV-anti-miR-331-3p, LV-anti-miR-331-3p+shMLLT10 or their negative control (LV-NC) were transfected into A549 cells; and LV-miR-331-3p, LV-miR-331-3p+LV-MLLT10 or their negative control (LV-NC) were transfected into H1650 cells to measure the effects of miR-331-3p and MLLT10 on the expression levels of MLLT10, E-cadherin and Vimentin, as well as tumor proliferation, migration, and invasion. The inhibitors and mimics of miR-331-3p, as well as lentiviruses mentioned above, were obtained from GenePharma (Shanghai, China).
RT-qPCR
Total RNA was collected from 26 paired tumor tissues and non-cancerous tissues, six types of cell lines without any transfection, A549 and H1944 cells transfected by inhibitor-miR-331-3p or inhibitor-NC, and also H1650 and H1299 cells transfected by mimic-miR-331-3p or mimic-NC, using TRIZOL reagent (Sigma, USA). The RNA was reverse transcribed to cDNA by using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA). The TaqMan MicroRNA PCR kit (Applied Biosystems, Foster City, CA), which contained primers of miR-331-3p and U6, was used to perform real-time qPCR with the help of the Applied Biosystems 7500 Realtime PCR System (Applied Biosystems, Foster City, CA, USA).
Luciferase Activity Assay
Wild type MLLT10 3ʹUTR or the mutant form of MLLT10 3ʹUTR (MLLT10 3ʹUTRmut) were inserted into pGL3 vector containing luciferase reporter gene and separately transfected into H1650 and H1299 cells placed in 6-well plates to carry out dual-luciferase activity assays. Following this, mimic-miR-331-3p or mimic-NC was added to the transfected H1650 and H1299 cells with the help of Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Forty-eight hours later, luciferase activity was measured in H1650 and H1299 cells with the help of a dual-luciferase reporter system (Promega Corporation, WI, USA). The sequences of MLLT10 3ʹUTR, MLLT10 3ʹUTRmut and pGL3 vector were bought from GenePharma (Shanghai, China).
Western Blot
A549 cells transfected with the lentiviruses LV-anti-miR-331-3p, LV-anti-miR-331-3p+shMLLT10, or LV-NC, and H1650 cells transfected with the lentiviruses LV-mimic-miR-331-3p, LV-mimic-331-3p+LV-MLLT10, or LV-NC, were lysed using ice-cold lysis buffer (Beyotime Biotechnology, Jiangsu, China). Proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the segregated proteins were transferred to polyvinylidene difluoride (PVDF) membranes. After blocking in 5% skimmed milk at room temperature for 1.5 h, the PVDF membranes were incubated with the specific primary antibodies (MLLT10, 1:1000, Abcam, Ambridge, UK, ab208016; E-cadherin, 1:10,000, Abcam, Ambridge, UK, ab40772; Vimentin, 1:2000, Abcam, Ambridge, UK, ab92547 and GADPH, 1:10,000, Abcam, Ambridge, UK, ab181602) at a controlled temperature of 4 °C. After 24 h, the membranes were washed with TBST three times and then incubated with the secondary antibody (1:20,000, Abcam, Ambridge, UK, ab205718) for two hours at room temperature to visualize the proteins. All antibodies used were purchased from Proteintech.
Cell Counting Kit-8 Assay
We applied the Cell Counting Kit-8 (CCK-8) assay to observe the effects of miR-331-3p and MLLT10 on tumor cell viability. Transfected cells were seeded into 96-well plates (3000 cells/well) and incubated at 37 °C with 5% CO2 atmosphere. During the five-day growth period, after 24 h of each addition of CCK8 solution (Beyotime, Hangzhou, China) to each well, cell numbers were counted using Thermo Scientific Fluoroskan Ascent (Thermo Scientific, USA) according to the optical density (OD) value measured at a wavelength of 450nm. Finally, a growth curve for cell growth in five days was generated.
Transwell Assay
Cell migration and cell invasion were both analyzed by the transwell assay. Transwell chambers (24-well plate, 8-µm pore size) were prepared for the following experiments. Transfected cells were washed with PBS two times, then cultured in RPMI-1640 medium without FBS for 24 h. 100ul cells with a density of 1×105 were seeded into the upper chambers. For the study of invasion assay, the uppers chambers were pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). The lower chambers, which were also called 24-well plates, were supplemented with 600ul RPMI-1640 medium with 10% FBS. After incubation for 24 h at 37 °C with 5% CO2, non-migrated cells or non-invaded cells in the upper chambers were wiped off gently using a cotton swab. Following this, the migrated or invaded cells were immobilized with 70% formaldehyde solution for 45 min, then stained with 0.1% crystal violet for half an hour. Eventually, five images of migrated or invaded cells, including the middle one and the peripheral four, were captured by a microscope.
Statistical Analysis
The data were represented as the mean ±standard deviation (SD), and were the average of three independent experiments. We carried out Student’s t-test and One-Way ANOVA analysis to measure the differences between quantitative variables. The above-mentioned statistical analyses were performed with the help of Prism (7.0) and R studio (3.6.1) software. Two-tailed P-value < 0.05 was considered to be statistically significant.
Results
miR-331-3p Was Down-Regulated in NSCLC
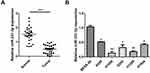
Our first objective was to investigate whether miR-331-3p was dysregulated in NSCLC. RT-qPCRs were carried out to measure the expression levels of miR-331-3p in NSCLC tumor tissues and cell lines. RT-qPCR results showed that the expression levels of miR-331-3p in 26 NSCLC tumor tissues were significantly lower than that in 26 paired non-cancerous tissues (Figure 1A). Also, the expression level of miR-331-3p in the five cell lines A549, H1650, H292, H1299, and H1944 was found to be lower than that found in BEAS-2b cell line, which is the normal human pulmonary epithelial cell line (Figure 1B). The above-mentioned positive and negative findings elucidated that miR-331-3p was downregulated in NSCLC, and was associated with NSCLC development to a large extent.
miR-331-3p Suppressed NSCLC Tumor Cell Proliferation
We used CCK-8 assay to identify the functional effects of miR-331-3p in NSCLC tumor cell proliferation. Firstly, RT-qPCR results confirmed that the expression levels of miR-331-3p were decreased in A549, H1944 and H1299 cells with the transfection of inhibitor-miR-331-3p (Figure 2A). Secondly, the results showed that the cell viabilities of A549, H1944 and H1299 cells transfected with inhibitor-miR-331-3p were higher than that of the cells transfected with inhibitor-NC (control group) from Day 3 to Day 5 correspondingly (Figure 2BD). On the other hand, the overexpression of miR-331-3p by the transfection of mimic-miR-331-3p in A549, H1650 and H1299 cells was confirmed by RT-qPCR results, as shown in Figure 2E. Subsequently, CCK-8 assay results elucidated that the cell viabilities of A549, H1650 and H1299 cells transfected with mimic-miR-331-3p were lower than that of the cells transfected with mimic-NC (control group) from Day 3 to Day 5 correspondingly (Figure 2FH). Overall, the results illustrated that the downregulation of miR-331-3p promoted the growth of A549, H1944 and H1299 cells, whereas the upregulation of miR-331-3p suppressed the growth of A549, H1650 and H1299 cells. Thus, by double verification, we have enumerated that miR-331-3p inhibits NSCLC tumor cell proliferation.
MLLT10 Was a Downstream Target Gene of miR-331-3p in the Development of NSCLC
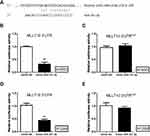
A dual-luciferase activity assay was performed to confirm that MLLT10 is a downstream target gene of miR-331-3p in the progression of NSCLC. Primarily, we observed that miR-331-3p could bind to a specific sequence in the 3ʹUTR region of MLLT10 mRNA (position 1470–1494) (Figure 3A). The luciferase activities of MLLT10 3ʹUTR in H1650 and H1299 cells transfected with mimic-miR-331-3p were lower than that in cells transfected with mimic-NC (Figure 3B and D). However, the luciferase activities for MLLT10 3ʹUTRmut in H1650 and H1299 cells transfected with mimic-miR-331-3p were identical with that in cells transfected with mimic-NC (Figure 3C and E). The above-mentioned findings showed that MLLT10 was a direct target of miR-331-3p in H1650 and H1299 cells, and thus we inferred that MLLT10 is a downstream target gene of miR-331-3p in the development of NSCLC.
MLLT10 Was Negatively Modulated by miR-331-3p in NSCLC
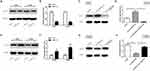
Western blot was used to explore the effects of miR-331-3p on MLLT10 expression. MLLT10 was downregulated in A549 and H1944 cells transfected with shMLLT10 (Figure 4A and B), and upregulated in H1650 and H1299 cells transfected with LV-MLLT10 (Figure 4E and F). Western blot results identified that the MLLT10 protein expression levels in A549 cells transfected with LV-anti-miR-331-3p were higher than that in A549 cells transfected with LV-NC or LV-anti-miR-331-3p+shMLLT10 (Figure 4C and D). Consistently, the MLLT10 protein expression levels in H1650 cells transfected with LV-miR-331-3p were lower than that in H1650 cells transfected with LV-NC or LV-miR-331-3p+LV-MLLT10 (Figure 4E and F). These results put together confirmed that MLLT10 was negatively modulated by miR-331-3p in NSCLC.
miR-331-3p Repressed Tumor Cell Proliferation, Metastasis and Invasion by Targeting MLLT10 in NSCLC
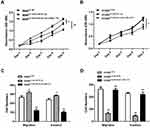
CCK-8 assay and transwell assay were used to investigate the functional roles of miR-331-3p and MLLT10 in the cellular processes of NSCLC. CCK-8 assay results revealed that the cell viability of A549 cells transfected with LV-anti-miR-331-3p was higher than that of cells transfected with LV-NC or LV-anti-miR-331-3p+shMLLT10 (Figure 5A). In addition, the cell viability of H1650 cells transfected with LV-miR-331-3p was evidently lower than that of cells transfected with LV-NC or LV-miR-331-3p+LV-MLLT10 (Figure 5B). Transwell assay showed that the migration and invasion cell numbers of A549 cells transfected with LV-anti-miR-331-3p were both significantly more than that of cells transfected with LV-NC or LV-anti-miR-331-3p+shMLLT10 (Figure 5C). In contrast, the migration and invasion cell numbers of A549 cells transfected with LV-miR-331-3p were both significantly less than that of cells transfected with LV-NC or LV-miR-331-3p+LV-MLLT10 (Figure 5D). Taking these results together, we propose that miR-331-3p represses tumor cell proliferation, migration, and invasion in NSCLC development. However, MLLT10 relieves these inhibitory effects of miR-331-3p on tumor cell proliferation, migration, and invasion in NSCLC development.
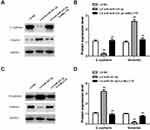
miR-331-3p/MLLT10 Axis Modulated the Expression of E-Cadherin and Vimentin in NSCLC
To further explore the mechanisms of metastasis in NSCLC tumor cells induced by miR-331-3p/MLLT10, E-cadherin and Vimentin expression levels were analyzed by Western blot. The Western blot results showed that E-cadherin expression levels in A549 cells transfected with LV-anti-miR-331-3p+shMLLT10 were significantly higher than that in A549 cells transfected with LV-anti-miR-331-3p alone, which served as the control group, whereas the Vimentin expression levels followed an opposite trend (Figure 6A and B). In addition, the E-cadherin expression levels in H1650 cells transfected with LV-miR-331-3p+LV-MLLT10 were significantly lower than that in H1650 cells transfected with LV-miR-331-3p alone, which also served as the control group. The Vimentin expression levels were consistent in following an opposite trend (Figure 6C and D). The suppression of the metastasis of A549 cells by the co-transfection of shMLLT10 correlated with the overexpression of E-cadherin and downregulation of Vimentin. On the other hand, the promotion of metastasis of H1650 cells by the co-transfection of LV-MLLT10 correlated with the downregulation of E-cadherin and overexpression of Vimentin. Thus, EMT-mediated metastasis induced by miR-331 and MLLT10 was verified by the above-mentioned experimental evidence.
Discussion
Our experimental results elucidated that miR-331-3p was downregulated in 26 NSCLC tumor tissues and five NSCLC cell lines, including A549, H1650, H292, H1299, and H1944. In addition, it was verified that MLLT10 was targeted directly and negatively modulated by miR-331-3p, which indicated that MLLT10 was a downstream target gene of miR-331-3p in NSCLC progression. Moreover, the overexpression of miR-331-3p inhibited, whereas its downregulation enhanced tumor cell proliferation, migration, and invasion in NSCLC. However, MLLT10 reversed the above-mentioned inhibitory effects of miR-331-3p. Thus, we inferred that miR-331-3p acts as an oncogene, whereas MLLT10 acts as an anti-oncogene in NSCLC. Consistently, we also found that the migration of NSCLC tumor cells induced by miR-331-3p and MLLT10 was EMT-mediated metastasis. These findings provided new insight into understanding NSCLC tumorigenesis and metastasis, and enumerated that miR-331-3p/MLLT10 axis and EMT may be useful targets for NSCLC diagnosis and therapy.
miR-331-3p has been validated to play different functional roles in several types of malignant carcinomas. For instance, miR-331-3p serves as a tumor suppressor in melanoma, colorectal cancer, cervical cancer, glioblastoma, and prostate cancer.25–29 On the other hand, miR-331-3p serves as a tumor promoter in pancreatic cancer and hepatocellular carcinoma.30–32 In the current study, miR-331-3p was shown to be a tumor suppressor in NSCLC development, which is consistent with previous studies. The functional role of miR-331-3p in a wide range of carcinomas, as well as other pathological diseases, needs to be explored further.
MLLT10 was a putative target gene of miR-331-3p as identified by bioinformatic approaches (TargetScan, http://www.targetscan.org/cgi-bin/targetscan/mamm_31/view_gene.cgi?taxid=9606andgs=MLLT10&members=miR-331). Our study illustrated not only that MLLT10 is a downstream target gene of miR-331-3p, but also that it is an oncogene in NSCLC cell proliferation, migration, and invasion. As far as we know, the research of MLLT10 has so far only focused on leukemia; thus, our study is the first to discover a correlation between MLLT10 and NSCLC. Recently, Jing et al proposed that MLLT10 conducted as a promoter in colorectal cell migration and invasion.33 This is another study to consider the potential effects of MLLT10 in malignant carcinomas, and its results provide evidence to validate our findings. The unknown role of MLLT10 in several diseases, especially human cancers, deserves further investigation.
To better understand the underlying mechanism of NSCLC tumor cell metastasis, and due to the ability of NSCLC tumor cells to easily transfer to another organ, the epithelial to mesenchymal transition (EMT) markers, E-cadherin and Vimentin, were examined. Our results revealed that miR-331-3p repressed the metastasis of NSCLC tumor cells by activating EMT. The tight correlation between miR-331-3p/MLLT10 and EMT-mediated metastasis was also found in hepatocellular carcinoma, pancreatic cancer, prostate cancer, and gastric cancer.30,32,34,35 MLLT10 was also validated to promote EMT-mediated metastasis in colorectal cancer.33 These reports were consistent with our finding that miR-331-3p/MLLT10 participated in the metastasis of NSCLC tumor cells via regulation of the expression of EMT markers.
Overall, our study proved that miR-331-3p is downregulated in NSCLC, and it inhibits NSCLC tumor cell proliferation, EMT-mediated metastasis, and invasion by targeting MLLT10. Although the detailed molecular mechanisms of miR-331-3p and MLLT10, the potential upstream gene of miR-331-3p and the downstream gene of MLLT10 in NSCLC, are not known and remain to be investigated in the future, however, the miR-331-3p/MLLT10 axis provides a novel clinical approach for NSCLC diagnosis, treatment and metastasis prevention.
Conclusions
Our study found miR-331-3p was down-regulated in NSCLC, and the down-regulation of miR-331-3p promoted NSCLC tumor cell proliferation, EMT-mediated metastasis and invasion by targeting MLLT10. miR-331-3p played a suppressor role in NSCLC tumorigenesis. Our findings highlighted miR-331-3p/MLLT10 axis could be useful as a clinical diagnostic marker and therapeutic target for NSCLC.
Disclosure
Qing-Qing Tian and Jing Xia are co first authors. The authors declare no conflicts of interest.
References
1. Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103(8):1144–1148. doi:10.1038/sj.bjc.6605901
2. Castro D, Moreira M, Gouveia AM, Pozza DH, De Mello RA. MicroRNAs in lung cancer. Oncotarget. 2017;8(46):81679–81685. doi:10.18632/oncotarget.20955
3. Lind H, Zienolddiny S, Ryberg D, Skaug V, Phillips DH, Haugen A. Interleukin 1 receptor antagonist gene polymorphism and risk of lung cancer: a possible interaction with polymorphisms in the interleukin 1 beta gene. Lung Cancer. 2005;50(3):285–290. doi:10.1016/j.lungcan.2005.07.003
4. Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378(9804):1727–1740. doi:10.1016/S0140-6736(10)62101-0
5. Barger JF, Nana-Sinkam SP. MicroRNA as tools and therapeutics in lung cancer. Respir Med. 2015;109(7):803–812. doi:10.1016/j.rmed.2015.02.006
6. Kang SM, Lee HJ. MicroRNAs in human lung cancer. Exp Biol Med. 2014;239(11):1505–1513. doi:10.1177/1535370214533887
7. Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi:10.1186/1476-4598-8-102
8. Zhang WC, Liu J, Xu X, Wang G. The role of microRNAs in lung cancer progression. Med Oncol. 2013;30(3):675. doi:10.1007/s12032-013-0675-8
9. Del Vescovo V, Grasso M, Barbareschi M, Denti MA. MicroRNAs as lung cancer biomarkers. World J Clin Oncol. 2014;5(4):604–620. doi:10.5306/wjco.v5.i4.604
10. Vannini I, Fanini F, Fabbri M. MicroRNAs as lung cancer biomarkers and key players in lung carcinogenesis. Clin Biochem. 2013;46(10–11):918–925. doi:10.1016/j.clinbiochem.2013.01.024
11. Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120(4):1298–1309. doi:10.1172/JCI39566
12. Liu L, Bi N, Wu L, et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16(1):50. doi:10.1186/s12943-017-0620-0
13. Geng GJ, Yang YT, Jiang J, Yu XY, Fa XE. MicroRNA-30a suppresses non-small-cell lung cancer by targeting Myb-related protein B. Exp Ther Med. 2018;15(2):1633–1639. doi:10.3892/etm.2017.5559
14. Liu S, Dong H, Dai H, Liu D, Wang Z. MicroRNA-216b regulated proliferation and invasion of non-small cell lung cancer by targeting SOX9. Oncol Lett. 2018;15(6):10077–10083. doi:10.3892/ol.2018.8573
15. Liu M, Zhou K, Cao Y. MicroRNA-944 affects cell growth by targeting EPHA7 in non-small cell lung cancer. Int J Mol Sci. 2016;17:10.
16. Han L, Chen W, Xia Y, et al. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am J Transl Res. 2018;10(4):1172–1183.
17. Lin X, Yang Z, Zhang P, Liu Y, Shao G. miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett. 2016;12(1):301–306. doi:10.3892/ol.2016.4577
18. Li S, Ma Y, Hou X, et al. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8(9):11854–11862.
19. Morerio C, Rapella A, Tassano E, Rosanda C, Panarello C. MLL-MLLT10 fusion gene in pediatric acute megakaryoblastic leukemia. Leuk Res. 2005;29(10):1223–1226. doi:10.1016/j.leukres.2005.03.008
20. Burillo-Sanz S, Morales-Camacho R, Vargas MT, Carrillo E, Perez-Soto I, Garcia-Lozano JR. A new rearrangement giving rise to a very rare MLL-MLLT10 fusion mRNA in an infant acute myeloid leukemia. Cancer Genet. 2015;208(3):101–102. doi:10.1016/j.cancergen.2014.10.006
21. Peterson JF, Sukov WR, Pitel BA, et al. Acute leukemias harboring KMT2A/MLLT10 fusion: a 10-year experience from a single genomics laboratory. Genes Chromosomes Cancer. 2019;58(8):567–577. doi:10.1002/gcc.22741
22. Naesens L, Devos H, Nollet F, Michaux L, Selleslag D. Mediastinal myeloid sarcoma with TP53 mutation preceding acute myeloid leukemia with a PICALM-MLLT10 fusion gene. Acta Haematol. 2018;140(2):97–104. doi:10.1159/000491596
23. Brandimarte L, Pierini V, Di Giacomo D, et al. New MLLT10 gene recombinations in pediatric T-acute lymphoblastic leukemia. Blood. 2013;121(25):5064–5067. doi:10.1182/blood-2013-02-487256
24. Othman MONEEBAK, Melo JB, Carreira IM, et al. MLLT10 and IL3 rearrangement together with a complex four-way translocation and trisomy 4 in a patient with early T-cell precursor acute lymphoblastic leukemia: A case report. Oncol Rep. 2015;33(2):625–630. doi:10.3892/or.2014.3624
25. Chen L, Ma G, Cao X, An X, Liu X. MicroRNA-331 inhibits proliferation and invasion of melanoma cells by targeting astrocyte-elevated gene-1. Oncol Res. 2018. doi:10.3727/096504018X15186047251584
26. Zhao D, Sui Y, Zheng X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep. 2016;35(2):1075–1082. doi:10.3892/or.2015.4450
27. Fujii T, Shimada K, Asano A, et al. MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int J Mol Sci. 2016;17(8):1351. doi:10.3390/ijms17081351
28. Epis MR, Giles KM, Candy PA, Webster RJ, Leedman PJ. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J Neurooncol. 2014;116(1):67–75. doi:10.1007/s11060-013-1271-7
29. Epis MR, Giles KM, Beveridge DJ, et al. miR-331-3p and Aurora Kinase inhibitor II co-treatment suppresses prostate cancer tumorigenesis and progression. Oncotarget. 2017;8(33):55116–55134. doi:10.18632/oncotarget.18664
30. Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2014;60(4):1251–1263. doi:10.1002/hep.27221
31. Cao Y, Chen J, Wang D, et al. Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget. 2015;6(35):38093–38106. doi:10.18632/oncotarget.5642
32. Chen X, Luo H, Li X, et al. miR-331-3p functions as an oncogene by targeting ST7L in pancreatic cancer. Carcinogenesis. 2018;39:1006–1015. doi:10.1093/carcin/bgy074
33. Jing X, Wu H, Cheng X, et al. MLLT10 promotes tumor migration, invasion, and metastasis in human colorectal cancer. Scand J Gastroenterol. 2018;53(8):964–971. doi:10.1080/00365521.2018.1481521
34. Fujii T, Shimada K, Tatsumi Y, Tanaka N, Fujimoto K, Konishi N. Syndecan-1 up-regulates microRNA-331-3p and mediates epithelial-to-mesenchymal transition in prostate cancer. Mol Carcinog. 2016;55(9):1378–1386. doi:10.1002/mc.22381
35. Zhang L, Song X, Chen X, et al. Circular RNA circcactin promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 Expression. Int J Biol Sci. 2019;15(5):1091–1103. doi:10.7150/ijbs.31533
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.