Back to Journals » OncoTargets and Therapy » Volume 11
miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/β-catenin signaling pathway in gastric carcinoma
Authors Xian X, Tang L, Wu C, Huang L
Received 28 July 2018
Accepted for publication 21 September 2018
Published 25 October 2018 Volume 2018:11 Pages 7503—7512
DOI https://doi.org/10.2147/OTT.S181706
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Xiangshu Xian,1,* Li Tang,2,* Chengrong Wu,1 Liuye Huang1
1Department of Gastroenterology, The Affiliated Yantai Yuhuangding Hospital of Qingdao University, Yantai, Shandong Province, People’s Republic of China; 2Department of Clinical Medicine, Qingdao University Medical College, Qingdao, Shandong Province, People’s Republic of China
*These authors contributed equally to this work
Background: Gastric cancer (GC) is the most common malignancy and third leading cause of cancer mortality worldwide. The identification of a sensitive biomarker as well as effective therapeutic targets for the treatment of GC is of critical importance. microRNAs play significant roles in the development of cancer and may serve as promising therapeutic targets.
Methods: The mRNA and protein expression of CB1R were studied both in GC cells and tissues. GC cell lines with specific gene overexpression and knockdown vectors were constructed. CCK-8 assay, matrigel invasion and colony formation assays were performed to evaluate the proliferation and invasion abilities. The binding and regulatory effects of miR-23b-3 and miR-130a-5p on CB1R mRNA were investigated using a luciferase reporter assay. Western blot analysis was performed to explore the potential interaction proteins of CB1R.
Results: In the present study, it was demonstrated that the cannabinoid receptor 1 (CB1R) was overexpressed, and miR-23b-3p and miR-130a-5p were downregulated, in GC cells. In addition, the results revealed that these effects are associated with malignant biological behaviors exhibited by GC cells. Furthermore, miR-23b-3p and miR-130a-5p may regulate CB1R expression via the Wnt/β-catenin signaling pathway.
Conclusion: Our results suggested dysregulation of CB1R expression is closely related to the malignant biological behavior of gastric cancer cells. miRNA/CB1R-based therapy may represent a promising therapeutic strategy for the clinical treatment of GC patients.
Keywords: miRNA, CB1R, Wnt, β-catenin, gastric carcinoma
Introduction
Gastric carcinoma (GC) is the fourth most common malignancy and the second most common cause of death of all malignancies worldwide.1,2 Despite declining trends worldwide, prevention of GC remains a priority in healthcare. Patients with GC exhibit symptoms during the advanced stages of the disease. The 5-year survival rate of patients with GC remains poor, varying between 10 and 30% in European countries.3 Recent studies have revealed that neoadjuvant therapy combined with surgery improves the 5-year progression-free rate compared with surgery alone.4 However, increased incidence of distant metastases and local recurrence post surgery continue to limit the prognosis of patients with GC.5 Identification of potential novel targets to reduce the recurrence of GC and prevent disease progression remains a significant clinical challenge.
Cannabinoid receptor 1 (CB1R) is a G-protein-coupled receptor activated by interaction with its endogenous ligands, anandamide and 2-arachidonoylglycerol, as well as a natural ligand isolated from the Cannabis sativa plant, delta-9-tetrahydrocannabinol.6,7 CB1 has been shown to activate ERK1/2 extracellular regulated kinases.8 Recent studies have revealed aberrant expression of CB1R is present in many tumors and is associated with clinical outcomes.9 It has been suggested that CB1R may represent a novel diagnostic and therapeutic target. However, few studies have investigated the association between CB1R and GC, and the role of CB1R in GC remains unclear.
MicroRNAs (miRNAs) are evolutionarily endogenous small non-coding RNAs that bind to specific complementary recognition sequences in the 3′-untranslated region (UTR) of the target mRNA sequence. Furthermore, miRNAs negatively regulate gene expression via either post-transcriptional inhibition or the targeting of mRNA for degradation in several diseases, such as cancer.10,11 Despite the already established important roles of microRNAs in various tumors, potential molecular associations between CB1R and microRNAs remain unknown.
In this study, it was demonstrated that CB1R is aberrantly overexpressed in GC cells and that the overexpression of CB1R promoted the viability and mobility of GC cells, predominantly via the Wnt/β-catenin signaling pathway. miR-23b-3p and miR-130a-5p may bind to the 3′-UTR of CB1R and regulate its expression. This study provides a novel insight into the association between CB1R and GC, and may promote the development of novel anticancer therapies for patients with GC.
Materials and methods
Patients and tissue specimens
Samples were collected from August 2018. Formalin-fixed paraffin-embedded tumor tissues and matched adjacent non-tumorous tissues were collected from six patients with GC who had previously undergone surgical resection as an initial treatment at our hospital. Diagnoses for included patients were confirmed via postoperative pathology investigation. The Hospital Research Ethics Committee approved the research protocol. Written informed consent and voluntary participation in the present study were obtained from all included patients prior to surgery.
Cell lines and culture
Human GC cell lines SGC-7901 and MKN-45, as well as the immortalized human gastric epithelial mucosa cell line GES-1, were purchased from the Cell Bank of Shanghai Institutes for Biological Sciences (Shanghai, China). All cell lines were cultured in DMEM supplemented with 10% fetal calf serum (HyClone; GE Healthcare Life Sciences, Marlborough, MA, USA), 100 U/mL penicillin and 100 μg/mL streptomycin, in a humidified atmosphere containing 5% CO2 at 37°C. Cells were cultured until a confluency of 80% was reached and were then digested using 0.25% trypsin and subsequently seeded into new plates at a required density.
A CB1R overexpression (CB1R OE) vector pCDH-CMV-CB1R-EF1-Puro and CB1R knockdown (CB1R KD) vectors, pLKO.1-CB1R shRNA1 and pLKO.1-CB1R shRNA2 were also constructed.
shRNA1 sequence:
5′-CCGGTGCTAATGTTTCCATAGTTTACTCGAGTAAACTATGGAAACATTAGCATTTTTG-3′ |
shRNA2 sequence:
5′-CCGGTAGTATCAGAGATGTCCATTTCTCGAGAAATGGACATCTCTGATACTATTTTTG-3′ |
Lentiviruses were packaged with the vectors and then SGC-7901 and MKN-45 cells were stably transfected to establish both CB1R overexpression and knockdown GC cell lines, respectively.
Cell counting kit-8 (CCK-8) assay
The effects of overexpression and knock down of CB1R on GC cell proliferation were investigated using Cell Counting Kit-8 (CCK-8) assays (Beyotime Institute of Biotechnology, Shanghai, China). SGC-7901 and MKN-45 cells were seeded in 96-well plates at a concentration of 5×103/100 μL per well and were then incubated for 48 hours. Following this, 10 μL of CCK8 was added and the plates were then incubated for 2 hours in the dark. The optical density (OD) at 450 nm was recorded using a microplate spectrophotometer.
Migration and invasion assays
The effects of overexpression and knock down of CB1R in SGC-7901 and MKN-45 cells on cell migration and invasion were analyzed using Transwell chambers (EMD Millipore, Billerica, MA, USA). Cells in the logarithmic growth phase were diluted with serum-free DMEM medium until a concentration of 5×104 cells/200 μL was reached. Two types of chambers were used: chambers with or without matrigel. A total of 200 μL of diluted cell suspension was added to each upper chamber, while 500 μL of medium containing 20% FBS was added to the lower chambers. After incubation for 48 hours, cells were fixed with 4% formaldehyde, stained using 10% crystal violet for 10–15 minute and then photographed using a microscope (200×).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells were extracted using TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), reverse-transcribed into cDNA and then amplified via qPCR (ABI 7500). The thermocycling conditions used for qPCR were as follows: 95°C for 2 minutes; followed by 45 cycles of 95°C for 5 seconds, 55°C for 20 seconds and 72°C, 30 seconds. A melting curve was established using the following conditions: 95°C for 15 seconds, 60°C for 1 minute and 95°C for 15 seconds. Assays were performed in triplicate. GAPDH was used as the control. The 2−ΔΔCT method was used to calculate the relative quantity of proteins. The primer sequences used were as follows:
Primers for CB1R:
Forward Sequence: 5′-CTGTTCCTCACAGCCATCGACA-3′ | |
Reverse Sequence: 5′-TGGCTATGGTCCACATCAGGCA-3′ |
Primers for GAPDH:
Forward Sequence: 5′-GTCTCCTCTGACTTCAACAGCG-3′ | |
Reverse Sequence: 5′-ACCACCCTGTTGCTGTAGCCAA-3′ |
Thermocycling conditions used for the qPCR amplification of microRNAs were as follows: 95°C for 10 minutes; followed by 40 cycles of 95°C for 10 seconds, 60°C for 20 seconds and 70°C for 30 seconds. In addition, a melting curve was also established.
Primers for microRNA:
> hsa-miR-23b-3p MIMAT0000418 | |
5′-AUCACAUUGCCAGGGAUUACCAC-3′ | |
Forward primer: 5′-ACACTCCAGCTGGGATCACATTGCCAGGGAT-3′; |
Reverse primer:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTGGTAAT-3′ | |
> hsa-miR-130a-5p MIMAT0004593 | |
5′-GCUCUUUUCACAUUGUGCUACU-3′ | |
Forward primer: 5′-ACACTCCAGCTGGGGCTCTTTTCACATTGT-3′; | |
Reverse primer: 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAG AGTAGCAC-3′ |
Primers for U6:
Forward Primer: 5′-CTCGCTTCGGCAGCACA-3′; | |
Reverse primer: 5′-AACGCTTCACGAATTTGCGT-3′. |
Western blot analysis
Proteins obtained from clinical specimens and cell lines were extracted using lysis buffer (Beyotime Institute of Biotechnology) and protease inhibitors (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Tissues and cell lysates were subjected to 10% PAGE and then transferred to PVDF membranes (PerkinElmer, Inc., Waltham, MA, USA). Membranes were blocked for 1 hour in 5% non-fat dry milk diluted with TBST (10 mM Tris–HCl and 0.05% Tween 20) and then immunoblotted with primary antibodies at 4°C overnight, followed by incubation with appropriate secondary antibodies at room temperature for 2 hours. The antibodies used were as follows: Anti-CB1R antibody (ab23703, dilute: 1:1,000, Abcam, Cambridge, UK), anti-Akt antibody (ab8805, dilute: 1:1,000, Abcam, UK), anti-p-Akt (phospho T308) antibody (ab8933, dilute: 1:1,000, Abcam, UK), anti-MMP-7 antibody (ab5706, dilute: 1:1,000, Abcam, UK), anti-β-catenin antibody (ab32572, dilute: 1:1,000, Abcam, UK), anti-c-Myc antibody (ab39688, dilute: 1:1,000, Abcam, UK), anti-cyclin D1 antibody (ab16663, dilute: 1:1,000, Abcam, UK), anti-mTOR antibody (ab2732, dilute: 1:1,000, Abcam, UK), anti-p-mTOR (phospho S2448) antibody (ab109268, dilute: 1:1,000, Abcam, UK), anti-P70S6K antibody (ab9366, dilute: 1:1,000, Abcam, UK), anti-p-P7036K (phospho T389) antibody (ab2571, dilute: 1:1,000, Abcam, UK), anti-GAPDH antibody (HRP) (ab9482, dilute: 1:5,000, Abcam, UK) and Goat Anti-Rabbit IgG H&L (HRP) (ab205718, dilute: 1:5,000, Abcam, UK). Western blotting was performed according to a general protocol and an ECL imaging kit was used to determine levels of chemiluminescent signaling.
Luciferase reporter gene assay
The binding and regulatory effects of miR-23b-3 and miR-130a-5p on CB1R mRNA were investigated using a luciferase reporter assay. Wild-type (WT) and mutant (MUT) 3′-UTR sequences of CB1R (5′-AAUGUGAA-3′ mutated to 5′-GGCACAGG-3′) were cloned into pGL-3M recombinant plasmids. The mimic and negative control of miR-23b-3p and miR-130a-5p were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and then separately combined with pGL-3M 3′-UTR WT or pGL-3M 3′-UTR MUT. Four different combinations (Mimic + WT, Mimic + MUT, control + WT and control + MUT) were transfected into human embryonic kidney HEK-293 cells. The relative activity of luciferase was detected using a Luciferase Assay System (Promega, Fitchburg, WI, USA).
Statistical analysis
All assays were performed in triplicate. Data analyses were performed using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA), and data were expressed as the mean ± SD. Quantitative experiments were analyzed using one-way ANOVA. P<0.05 was considered to indicate a statistically significant difference. Statistical graphs were established using Graphpad Prism five software.
Results
CB1R is highly expressed in gastric cancer cells
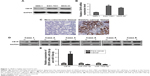
RT-qPCR assays were performed to investigate the expression levels of CB1R in SGC-7901 and MKN-45 human GC cell lines, as well as in the human gastric epithelial mucosa cell line, GES-1. The results demonstrated that the expression level of CB1 mRNA was 2.5–3.5 times higher in GC cells compared with normal gastric epithelial cells (P<0.05) (Figure 1B). Additionally, Western blotting was performed to evaluate the expression level of CB1R protein, and the results showed that the expression levels of CB1R were increased in both SGC-7901 and MKN-45 cells compared with the level in GES-1 cells (Figure 1A), which was in accordance with the RT-qPCR results. Meanwhile, the expression of CB1R was higher in GC tissues compared with corresponding non-cancerous tissues (Figure 1C). Levels of CB1R mRNA and protein expression were determined in GC tissues and non-cancerous tissues via RT-qPCR and Western blot assays. As presented in Figure 1D and E, the relative expression levels of CB1R were increased in GC tissues compared with non-cancerous tissues (n=6, P<0.05). Furthermore, the expression levels of miR-23b-3p and miR-130a-5p were downregulated in tumors compared with corresponding non-cancerous tissues (Figure 1E).
Increased expression of CB1R promotes the proliferation and invasion of GC cells
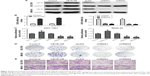
The role of CB1R in GC has not previously been investigated. To further evaluate the effect of CB1R expression on malignant biological behaviors in GC, such as cell proliferation, migration and invasion ability, stable CB1R overexpression (OE) and knockdown (KD) GC cell lines were established via lentiviral transfection. Following the confirmation of CB1R expression levels in OE and KD cell lines via RT-qPCR and Western blotting (Figure 2A and B), CCK-8, colony formation, transwell and wound-healing assays were performed to examine the biological characteristics of the stably transfected cells to determine the effects of CB1R on gastric cell proliferation, invasion and migration. The results revealed that overexpression of CB1R promoted the proliferation and clonal formation of GC cells (Figure 2C and D). The results of the transwell assays demonstrated that overexpression of CB1R enhanced the invasion and migration ability of GC cells (Figure 2E); however, CB1R KD SGC-7901 and CB1R KD MKN-45 cells exhibited opposite results (Figure 2C–E). Therefore, enhanced expression of CB1R is closely associated with malignant biological behaviors exhibited by GC cells.
Expression levels of miR-23b-3p and miR-130a-5p are significantly suppressed in GC cells
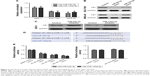
Potential microRNAs that may be involved in the regulation of CB1R expression were predicted using TargetScanHuman 7.1 software (http://www.targetscan.org/vert_71/). Following this, candidate microRNAs were analyzed using miRCancer (http://mircancer.ecu.edu), and the results revealed that the expression levels of miR-23b-3p and miR-130a-5p were downregulated. In addition, the relative expression levels of miR-23b-3p and miR-130a-5p in SGC-7901, MKN-45 and GES-1 cells were investigated, and the results demonstrated that the expression levels of miR-23b-3p and miR-130a-5p in the SGC-7901 and MKN-45 gastric cancer cell lines were suppressed compared with the normal gastric epithelial mucosa cell line, GES-1 (Figure 3A).
Furthermore, the expression levels of miR-23b-3p in SGC-7901 and MKN-45 cells were 61% and 43% compared with that in GES-1 cells, respectively. The expression level of miR-130a-5p in SGC-7901 and MKN-45 cells was 50% and 52% compared with that in GES-1 cells (Figure 3A).
microRNAs regulate CB1R expression via binding to the 3′-UTR region of CB1R
SGC-7901 and MKN-45 cells transiently transfected with miR-23b-3p mimics (mimic1) and miR-130a-5p mimics (mimic2) were subjected to Western blotting, and the results revealed that both mimics inhibited CB1R expression (Figure 3B). Treatment with microRNA inhibitors upregulated the expression of CB1R in normal GES-1 gastric epithelial cells (Figure 3C). Therefore, the results suggested that CB1R represents a target gene of miR-23b-3p and miR-130a-5p.
Regions of CB1R mRNA that represent binding sites for miR-23b-3p and miR-130a-5p were predicted by TargetScanHuman 7.1 (http://www.targetscan.org/vert_71/), and the results are presented in Figure 3D. Luciferase reporter gene assays were performed to verify the binding sites and regulatory effects of miR-23b-3p and miR-130a-5p on CB1R mRNA. The results indicated that human embryonic kidney HEK-293 cells co-transfected with miR-23b-3p/hsa-miR-130a-5p mimics and pGL-3M 3′-UTR WT exhibited significantly decreased luciferase activity compared with the controls, which suggested that both miR-23b-3p and miR-130a-5p can bind to the wild-type 3′-UTR sequence of CB1R and subsequently downregulate the expression of CB1R (Figure 3E). However, the luciferase activities exhibited by cells co-transfected with miR-23b-3p/miR-130a-5p mimics and pGL-3M 3′-UTR MUT were similar compared with the control, which suggested that miR-23b-3p and miR-130a-5p were unable to bind to the mutated 3′-UTR sequence of CB1R and subsequently regulate the expression of CB1R (Figure 3E). Therefore, the results indicated that miR-23b-3p and miR-130a-5p may bind to the 3′-UTR of CB1R at the sites identified in Figure 3D and subsequently regulate the expression of CB1R.
Increased expression of CB1R promotes the proliferation, migration and invasion of GC cells by activating the Wnt/β-catenin pathway
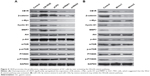
The Wnt/β-catenin and PI3K/Akt/mTOR/P70S6K pathways have important roles in cancer cell proliferation, apoptosis, migration and invasion. The expression levels of important molecules in two signaling pathways were detected in CB1R OE and KD SGC-7901 and MKN-45 cells. The results demonstrated that overexpression of CB1R significantly upregulated the expression of β-catenin, c-Myc, cyclin D1 and MMP7 in SGC-7901 cells, which suggested that the Wnt/β-catenin pathway was activated. However, the Wnt/β-catenin pathway was revealed to be markedly inhibited in CB1R KD cells (Figure 4A).
However, no marked changes in p-Akt, p-mTOR and p-P7036K levels were observed in either CB1R OE cells or CB1R KD cells, which indicated that CB1R promoted the proliferation, migration and invasion of GC cells via the Wnt/β-catenin pathway, but not the PI3K/Akt/mTOR/P70S6K pathway (Figure 4A). The results suggest that the increased expression of CB1R promoted the occurrence and development of GC.
In addition, SGC-7901 cells transiently transfected with miR-23b-3p mimics (mimic1) and miR-130a-5p mimics (mimic2) were subjected to Western blotting analysis, and the results revealed that both mimics inhibited the Wnt/β-catenin pathway, and similar results were exhibited by CB1R KD cells (Figure 4B). However, the PI3K/Akt/mTOR/P70S6K pathway in these transiently transfected cells was also slightly inhibited, which suggested that miR-23b-3p mimics (mimic1) and hsa-miR-130a-5p mimics (mimic2) may also target numerous genes involved in the PI3K/Akt/mTOR/P70S6K pathway.
Discussion
Gastric carcinoma is a malignant disease with a generally poor long-term prognosis. Numerous studies have led to the identification of various biomarkers and signaling pathways associated with the prognosis of patients with GC, including CDX2, MUC13, GP87 and COX-2.12–15 A sensitive biomarker that is able to predict metastatic potential in individual cases of GC would be of clinical significance; however, no such biomarkers have been clinically applied thus far. Despite cannabinoids being associated with antineoplastic activity in a number of cancer cell types, the effect of cannabinoids on GC cells has not been clarified. Xian et al16 suggested that cannabinoids may represent a novel GC therapy and that the WIN 55,212–2 agonist inhibits the proliferation of human GC cells in a dose-dependent manner; however, this effect may be partially mediated by CB1R. In the present study, the results revealed that CB1R is overexpressed in GC cells. Furthermore, stable CB1R OE and KD GC cell lines were established to investigate malignant biological behaviors associated with GC cells. The results demonstrated that overexpression of CB1R promoted the proliferation, clonal formation, migration and invasion of GC cells; however, these effects were attenuated via transfection with CB1R-shRNA. miRNAs can function both as oncogenes and tumor suppressors during tumor progression. To the best of our knowledge, potential associations between the cannabinoid receptor and miRNAs in GC have not yet been studied. In the present study, potential microRNAs (miR-23b-3p and miR-130a-5p) that may be involved in the regulation of CB1R expression were predicted using TargetScanHuman 7.1 software (http://www.targetscan.org/vert_71/). The results revealed that the expression levels of miR-23b-3p and miR-130a-5p in GC cells were decreased compared with GES-1 cells. Transfection with mimics decreased the protein expression of CB1R in GC cells. The results of the luciferase reporter gene assay verified that miR-23b-3p and miR-130a-5p can bind to the 3′-UTR of CB1R and regulate the expression of CB1R.
Numerous studies have investigated the molecular mechanisms underlying the induction of primary tumor development; however, signaling pathways associated with cancer metastasis remain unclear. The Wnt/β-catenin and PI3K/Akt/mTOR/P70S6K pathways have important roles in cancer cell proliferation, apoptosis, migration, and invasion.17–20 Recent studies have suggested that miRNAs either function as tumor suppressors or oncogenes in the post-transcriptional regulation of tumor development and metastasis by regulating several signaling pathways, such as the Wnt/β-catenin, Notch, TGF-β and PI3K/Akt pathways.21,22 The results of the present study revealed that enhanced expression of CB1R promoted the occurrence and development of GC, predominantly via the Wnt/β-catenin signaling pathway, but not the PI3K/Akt/mTOR/P70S6K pathway. However, the PI3K/Akt/mTOR/P70S6K pathway in transiently transfected GC cells was also slightly inhibited, which suggested that miR-23b-3p mimics (mimic1) and miR-130a-5p mimics (mimic2) may also target numerous genes associated with the PI3K/Akt/mTOR/P70S6K pathway. Thus, multiple mechanisms are likely to be associated with CB1R-induced tumor progression. The results of the present study suggested that CB1R-induced tumor progression may be associated with the regulation of CB1R by miR-23b-3p and miR-130a-5p via the Wnt/β-catenin signaling pathway.
Conclusion
In conclusion, the present study suggested that the CB1R receptor is associated with the promotion of malignant biological phenotypes in GC cells, and provides additional information regarding the complex interaction between CB1R and tumor progression. The regulation of CB1R via binding with miR-23b-3p and miR-130a-5p in the Wnt/β-catenin signaling pathway may have an important role in tumor progression. However, complex crosstalk in the tumor microenvironment may additionally contribute to the progression and metastasis of tumors. Moreover, miRNAs may simultaneously regulate several genes and modify their functions as tumor suppressors and oncogenes. Finally, miRNAs/CB1R-based therapy may represent a promising strategy for the treatment of patients with GC; however, further investigations are necessary to verify the results of the present study.
Acknowledgments
This work was supported by grants from the Key research and development plan of Shangdong Province (No 2017GSF218020), the Key research and development plan of Yantai (No 2017YD011) and the National Natural Science Foundation of China (No 81402488).
Disclosure
The authors report no conflicts of interest in this work.
References
Wright NA, Poulsom R, Stamp G, et al. Trefoil peptide gene expression in gastrointestinal epithelial cells in inflammatory bowel disease. Gastroenterology. 1993;104(1):12–20. | ||
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. | ||
Cunningham D, Allum WH, Stenning SP, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355(1):11–20. | ||
D’Angelica M, Gonen M, Brennan MF, Turnbull AD, Bains M, Karpeh MS. Patterns of initial recurrence in completely resected gastric adenocarcinoma. Ann Surg. 2004;240(5):808–816. | ||
Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346(6284):561–564. | ||
Gérard CM, Mollereau C, Vassart G, Parmentier M. Molecular cloning of a human cannabinoid receptor which is also expressed in testis. Biochem J. 1991;279(Pt 1):129–134. | ||
Asimaki O, Mangoura D. Cannabinoid receptor 1 induces a biphasic ERK activation via multiprotein signaling complex formation of proximal kinases PKCε, Src, and Fyn in primary neurons. Neurochem Int. 2011;58(2):135–144. | ||
Chakravarti B, Ravi J, Ganju RK. Cannabinoids as therapeutic agents in cancer: current status and future implications. Oncotarget. 2014;5(15):5852–5872. | ||
Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16(3):223–229. | ||
Lai EC. miRNAs: whys and wherefores of miRNA-mediated regulation. Curr Biol. 2005;15(12):R458–R460. | ||
Saito M, Okayama H, Saito K, et al. CDX2 is involved in microRNA-associated inflammatory carcinogenesis in gastric cancer. Oncol Lett. 2017;14(5):6184–6190. | ||
He L, Qu L, Wei L, Chen Y, Suo J. Reduction of miR-132-3p contributes to gastric cancer proliferation by targeting MUC13. Mol Med Rep. 2017;15(5):3055–3061. | ||
Qiao SX, Yuan M, Liu YL, Lin XS, Zhang XP, Tobi M. Detection of gastric cancer and premalignant lesions by novel marker glycoprotein 87 using monoclonal antibody Adnab-9. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1095–1099. | ||
Ye Y, Liu M, Yuan H, et al. COX-2 regulates Snail expression in gastric cancer via the Notch1 signaling pathway. Int J Mol Med. 2017;40(2):512–522. | ||
Xian XS, Park H, Cho YK, et al. Effect of a synthetic cannabinoid agonist on the proliferation and invasion of gastric cancer cells. J Cell Biochem. 2010;110(2):321–332. | ||
Sebio A, Kahn M, Lenz HJ. The potential of targeting Wnt/β-catenin in colon cancer. Expert Opin Ther Targets. 2014;18(6):611–615. | ||
Suwala AK, Koch K, Rios DH, et al. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. Oncotarget. 2018;9(32):22703–22716. | ||
Zhu J, Yao J, Huang R, Wang Y, Jia M, Huang Y. Ghrelin promotes human non-small cell lung cancer A549 cell proliferation through PI3K/Akt/mTOR/P70S6K and ERK signaling pathways. Biochem Biophys Res Commun. 2018;498(3):616–620. | ||
Wang H, Duan L, Zou Z, et al. Activation of the PI3K/Akt/mTOR/p70S6K pathway is involved in S100A4-induced viability and migration in colorectal cancer cells. Int J Med Sci. 2014;11(8):841–849. | ||
Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer. 2015;51(12):1638–1649. | ||
Moyret-Lalle C, Ruiz E, Puisieux A. Epithelial-mesenchymal transition transcription factors and miRNAs: “Plastic surgeons” of breast cancer. World J Clin Oncol. 2014;5(3):311–322. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.