Back to Journals » Cancer Management and Research » Volume 12
miR-105 Promotes the Progression and Predicts the Prognosis for Oral Squamous Cell Carcinoma (OSCC)
Received 23 May 2020
Accepted for publication 4 September 2020
Published 10 November 2020 Volume 2020:12 Pages 11491—11499
DOI https://doi.org/10.2147/CMAR.S262442
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Hong-Yu Zhang,1 Jian-Hui Ma2
1Department of Oral and Maxillofacial Surgery, Affiliated Stomatological Hospital of Jiamusi University, Jiamusi, Heilongjiang 154004, People’s Republic of China; 2Department of Stomatology, The 80th Group Army Hospital of the PLA, Weifang, Shandong 261021, People’s Republic of China
Correspondence: Jian-Hui Ma
Department of Stomatology, The 80th Group Army Hospital of the PLA, No. 256, Beigong West Street, Weifang, Shandong 261021, People’s Republic of China
Email [email protected]
Background: miRNA-105 has been reported in a vast number of malignancies, including hepatocellular carcinoma and colorectal, esophageal, breast and non-small lung cancers. Still, the biological role of miR-105 remains mostly uncovered in oral squamous cell carcinoma (OSCC).
Methods: miR-105 expression in OSCC tissues and cell lines was detected by qRT-PCR. Survival analysis was performed using the Kaplan–Meier method, while the prognostic significance of miR-105 was evaluated by Cox regression analysis with a cohort of 90 OSCC patients. The effects of miR-105 on the proliferation of tumor cells were analyzed by CCK-8 assay and crystal violet staining, while cell invasion was assessed by transwell assays.
Results: Our current work indicates that miR-105 was upregulated in human OSCC tissues and cell lines. Moreover, miR-105 expression was closely associated with tumor size as well as clinical and differentiation stages. Notably, an elevated expression of miR-105 may predict some poor clinical prognosis in OSCC patients. Furthermore, miR-105 overexpression can significantly promote the proliferation and invasion of OSCC cells, whereas downregulation of miR-105 inhibits these cellular events.
Conclusion: This study demonstrates that miR-105 can promote the proliferation and invasion of OSCC cells. High expression of miR-105 predicts poor prognosis for OSCC and, therefore, it may represent a prognostic biomarker and putative therapeutic target for patients affected by OSCC.
Keywords: miR-105, cancer progression, prognosis, biomarker, OSCC
Introduction
Oral squamous cell carcinoma (OSCC) is characterized as carcinoma derived from the epithelium lining the oral cavity, which includes tongue, cheek, gingiva and floor of mouth.1 OSCC is considered the sixth most common type of epithelial neoplasm that develops in the head and neck region, accounting for ~4% of all cancer cases.2 The incidence and mortality due to this disease vary geographically. Over 300,000 new cases worldwide occur every year, where oral cancer is responsible for more than 140,000 deaths annually.3,4 Despite the clinical use of multiple therapeutic approaches, including surgery, radiotherapy, targeted therapy, and immunotherapy, the prognosis for OSCC remains mostly suboptimal. In fact, over half of the affected patients have an overall survival time lower than 5 years.5 Thus, identifying novel prognostic biomarkers and therapeutic targets is crucial for the diagnosis and therapy for OSCC patients.
MicroRNAs (miRNAs) correspond to a subset of small non-coding RNAs with an average length of 20–25 nucleotides. In recent years, numerous studies have shown that miRNAs participate in a wide range of biological processes, including cell proliferation, migration, invasion, and apoptosis, eventually providing roles as oncogenes or tumor suppressor genes in multiple types of cancer.6–8 The diagnostic and prognostic roles of miRNAs have been increasingly elucidated in OSCC patients, thus providing novel insights into alternate treatment modalities. MiRNA-105 is encoded by a gene located in the chromosome Xq28.9 Interestingly, the roles of miRNA-105 have been defined in a large variety of cancers. Indeed, the oncogenic role of miR‐105 has been confirmed in many malignancies including colorectal,10 esophageal,11 breast12 and non-small lung cancer.13 In contrast, multiple studies have proposed a tumor suppression role for miR-105 in other types of cancer such as hepatocellular carcinoma,14 glioma,15 gastric16 and prostate cancer.17 However, the role and function of miRNA-105 remain poorly characterized in OSCC.
Accordingly, the present study aims to detect the prognostic and biological functions of miR-105 during OSCC progression. For this, we initially verified the expression of miR-105 in human OSCC tissues and cell lines. Thereafter, we explored the clinical prognostic significance of miR-105 in OSCC patients, to focus on whether its expression may predict the disease prognosis. Additionally, the biological functions of miR-105 were also investigated during the progression of OSCC cells in vitro.
Patients and Methods
Patient Samples and Follow-Up
A total of 90 OSCC patients, hospitalized at the Affiliated Stomatological Hospital of Jiamusi University (Heilongjiang, China) between January 2015 and December 2017, were enrolled in this study. All patients underwent routine surgery without receiving any previous treatments (ie, chemotherapy or radiation). OSCC tissues and non-cancerous counterparts were collected and stored at −80°C until further RNA extraction. Patients were followed-up once every 3–4 months until being deceased or dropped out from the follow-up program. Clinicopathological characteristics, including gender, age, smoking habits, tumor size, clinical stage, cervical nodal invasion, tumor differentiation, and tumor location, as well as survival information, were properly recorded. The histopathological differentiation stage of the tumor as well as the clinical stage of respective patients was evaluated according to the WHO grading classification and AJCC 8th staging system, respectively. Regular physical examinations were conducted for all patients to determine any disease recurrence and/or the status of cervical node invasion. Overall survival (OS) was defined as the interval between the date of surgery and the death date (or last follow-up). Disease-free survival (DFS) was defined as the interval between the date of surgery and the date of death, recurrence, or the last clinical follow-up. This study was censored on January 2020. At a median follow-up of 28 months (range of 15–60 months), 38 patients were deceased while 5 patients failed to follow-up. All informed consents were obtained from patients included in this study. This work was approved by the Ethics Committee of the Affiliated Stomatological Hospital of Jiamusi University (ASHJU-20,200,013) and conducted in accordance with the Declaration of Helsinki.
MiRNA Extraction and Quantitative Reverse Transcription‑Polymerase Chain Reaction (qRT-PCR)
The overall content of miRNAs from both clinical and cell line samples were extracted according to the manufacturer’s protocol (miRNeasy Mini kit, Qiagen). MiR-105-specific primers and probe (TaqMan MicroRNA Assay Kit, Applied Biosystems) were used to evaluate its expression levels using an ABI 7300 system (Applied Biosystems). The thermal cycling conditions were as it follows: 40 cycles of 95°C for 20 secs, 55°C for 30 secs, and 72°C for 30 secs. The relative miR-105 expression was normalized according to U6 miRNA levels, using the 2−∆∆Ct method.
Cell Culture
Human OSCC cell lines (Cal27, SCC9, and HSU3) and normal oral keratinocytes (HOK) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Sangong Biotech), and incubated at 37 ◦C in a humidified atmosphere of 5% CO2.
Cell Transfection
MiR-105 mimics and inhibitor, as well as their corresponding negative controls (NCs), were obtained from GenePharma (Shanghai, China). OSCC cells were seeded in 6-well plates (3×105 per well) and cultured for 24 hrs before transfection. Cells were further transfected, according to the manufacturer’s protocol, using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 hrs of transfection, cells were harvested for subsequent experiments.
Cell Counting Kit-8 (CCK-8) Assay
Cells at the density of 1×104 per well were seeded, in triplicates, into 96-well plates. A volume of 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan) was added to the medium at 0, 24, 48, and 72 hrs of cell culture, and then incubated at 37 °C for additional 2 hours. Absorbance was measured at 450nm using an automatic microplate reader.
Crystal Violet Staining
Cells were seeded into 6-well plates at a density of 1000 cells per well. Cells were cultured in a medium containing 10% FBS, which was replaced every three days. Two weeks later, medium was removed and cells were then fixed and stained with crystal violet solution (1 mL 0.5% crystal violet solution in 20% methanol). Ten minutes later, fixed cells were washed with phosphate-buffered saline (PBS) and photographed. Absorbance was measured at 570 nm using an automatic microplate reader.
Transwell Assay
To estimate the invasion capability of OSCC cells, transwell assays were performed using a 24-well Transwell chamber (8.0 μm, Corning). For this, a total of 150 μL cell suspension (5 × 104 cells) were seeded in respective upper chambers containing FBS-free DMEM. Lower chambers were loaded with DMEM supplemented with 10% FBS. Membranes were pre-coated with 50 µL Matrigel (BD Biosciences). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. After 24-hr incubation, invasive cells were fixed with 100% methanol and then stained with 0.1% crystal violet for 10 mins at room temperature. Five fields were randomly selected for cell counting, using a light microscope (Olympus Corporation) at ×200 magnification.
Statistical Analysis
All data are expressed as mean ± standard deviation. Statistical analyses were performed with GraphPad Prism 5.0 (GraphPad Software Inc., CA, USA) and SPSS 20.0 (IBM SPSS Inc., Chicago, IL, USA). Student’s t-test was pursued for the analysis of different groups. The clinicopathological parameters were presented as categorical data and then compared by chi-square test or Fisher’s exact test. Kaplan–Meier curves were generated for OS and DFS, and the respective differences were analyzed by Log rank test. Univariate and multivariate Cox regression analyses with stepwise selection were performed to detect independent predictors of OS and DFS. Significant prognostic factors from the univariate analysis (P< 0.05) were subjected to multivariate analysis using the Cox proportional hazards regression model. A p-value of less than 0.05 denoted a statistically significant difference.
Results
MiR-0105 is Upregulated in OSCC Tissues and Related Cell Lines
The expression of miR-105 was examined in 90 pairs of OSCC tissues and respective non-cancerous counterparts by qRT-PCR. Our results showed that the expression of miR-105 was significantly upregulated in tumor tissues when compared to the non-cancerous controls (P < 0.001, Figure 1A). The expression of miR-105 was also detected in several OSCC cell lines as well as normal oral keratinocytes. As shown in Figure 1B, we observed that miR-105 expression was significantly higher in OSCC cell lines (Cal27, SCC9, and HSU3) when compared to normal oral keratinocytes (HOK) (P < 0.001). According to the median value of miR-105 expression in tumor tissues, OSCC patients were divided into two groups: miR-105 low (below the median, n=45) and miR-105 high (above the median, n=45).
Clinical Significance of miR-0105 Expression in OSCC Patients
The relationship between miR-105 expression and the clinicopathologic characteristics of OSCC was evaluated in our selected group of patients. As shown in Table 1, patients with high miR-105 expression presented a higher proportion of T3-T4 tumors than those with low expression of miR-105 (P = 0.020). Consistently, the number of patients with high miR-105 expression was also higher in clinical stages III-IV (P = 0.006). Moreover, patients with poor differentiation stage were more common among the miR-105 high expression than the low expression group (P = 0.036). However, no significant difference between the two groups was observed in regard to age, gender, smoking status, cervical nodal invasion, and tumor location.
 |
Table 1 miR-105 Expression and Clinical Characteristics in 90 Cases of OSCC |
Kaplan–Meier curves showed that OSCC patients with low miR-105 expression presented better OS (Figure 2A; P = 0.002) and DFS (Figure 2B; P = 0.03) when compared to those with high miR-105 expression. Multivariate Cox regression analysis was also performed to further explore the prognostic factors related to OS and DFS. Therefore, some prognostic factors for OS (Table 2) were presently found. These factors were (i) tumor size [hazard ratio (HR): 2.022, 95% confidence interval (CI): 1.108–3.690, P = 0.022]; (ii) cervical nodal invasion (HR: 2.753, 95% CI: 1.456–5.205, P = 0.002); and (iii) miR-105 expression (HR: 2.038, 95% CI: 1.145–3.628, P = 0.015). Coincidentally, the prognostic factors for DFS were (i) tumor size (HR: 2.371, 95% CI: 1.229–4.576, P = 0.010); (ii) cervical nodal invasion (HR: 2.200, 95% CI: 1.092–4.431, P = 0.027); and (iii) miR-105 expression (HR: 2.371, 95% CI: 1.176–4.777, P = 0.016) (Table 3). Taken together, these findings indicate that is upregulated in OSCC tissues, and higher miR-105 expression correlates with a poor prognosis in OSCC patients.
 |
Table 2 Univariate and Multivariate Analyses of Prognostic Factors Associated with Overall Survival |
 |
Table 3 Univariate and Multivariate Analyses of Prognostic Factors Associated with Disease-Free Survival |
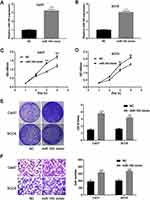
Overexpression of miR-105 Promotes the Proliferation and Invasion of OSCC Cells
Based on the above results, further experiments were conducted to explore the biological functions of miR-105 during OSCC progression. Hence, we overexpressed miR-105 in Cal27 and SCC9 cell lines using miR-105 mimics, and then examined the effects on cell proliferation and invasion. Based on qRT-PCR analysis, we were able to confirm that miR-105 mimics can significantly increase miR-105 expression in these selected cell lines (P < 0.001, Figure 3A and B). CCK-8 assays showed that the absorbance values obtained from Cal27 and SCC9 cells, after 3 and 4 days of transfection with miR-105 mimics, were significantly higher than those from untreated cells (P < 0.01, Figure 3C and D). Moreover, crystal violet staining indicated that the overexpression of miR-105 can also significantly promote the proliferation of Cal27 and SCC9 cells (P < 0.001, Figure 3E). Transwell assays were further performed to explore the invasion capability of OSCC cells. As shown in Figure 3F, miR-105 overexpression was able to highly promote the invasion of Cal27 and SCC9 cells (all P < 0.001).
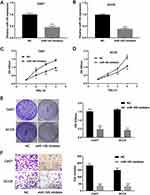
Downregulation of miR-105 Inhibits the Proliferation and Invasion of OSCC Cells
Specific inhibitors were further utilized to downregulate the expression of miR-105 in vitro. The efficacy of this inhibition was confirmed by qRT-PCR (P < 0.001, Figure 4A and B). CCK-8 assays and crystal violet staining showed that miR-105 downregulation significantly inhibited the proliferation of both Cal27 and SCC9 cells (P < 0.05, Figure 4C–E). Coincidentally, transwell assays also demonstrated that the downregulation of miR-105 may significantly inhibit the invasion of Cal27 and SCC9 cells (P < 0.001, Figure 4F).
Discussion
A variety of miRNAs have been currently identified as putative diagnostic or prognostic biomarkers by exerting fundamental roles in OSCC progression.18–20 For instance, miR-361-3p is upregulated in OSCC tissues and thus facilitates the growth of human OSCC cells, both in vitro and in vivo.21 Moreover, salivary exosomal miR-24-3p has been also reported to maintain the growth of OSCC by targeting PER1 and, therefore, to act as a potential diagnostic biomarker.22 Chen and colleagues have reported that miR-1254 may restrain the progression of OSCC by downregulating CD36, which may also represent an effective treatment strategy for affected patients.23
In the present study, we have documented that miR-105 is upregulated in human OSCC tissues and cell lines when compared with non-cancerous tissues and HOK cells, respectively. We have demonstrated that miR-105 expression is closely associated with tumor size as well as clinical and differentiation stages. Notably, higher miR-105 expression can serve as a predictor of poor clinical prognosis in OSCC patients. These findings suggest that miR-105 might act as an independent prognostic biomarker and also as an oncogene in OSCC. Subsequently, we modulated miR-105 expression by overexpressing and downregulating it in two OSCC cell lines using miR-105 mimics and inhibitors, respectively. Next, the effects of miR-105 towards the proliferation and invasion of OSCC cells were explored. As a result, we found that overexpressing miR-105 could significantly promote the proliferation and invasion of OSCC cells, whereas downregulating it inhibited these cellular effects.
The oncogenic role of miR-105 has been confirmed in several types of malignancies, including colorectal, esophageal, breast and non-small lung cancers. Shen and colleagues have verified that miR-105 overexpression can be associated with an aggressive phenotype in colorectal cancer, being necessary for the TNF-α-induced epithelial–mesenchymal transition.10 Gao and colleagues have reported that a higher miR-105 expression may predict poorer clinical prognosis in esophageal cancer patients.11 Consistent with our findings, miR-105 can also promote the proliferation, migration, and invasion of esophageal cancer cells in vitro.11 Moreover, miR-105 has been shown to promote stemness, chemoresistance, and metastasis, thus serving as a putative diagnostic biomarker in triple-negative breast cancer.12 Overexpression of miR‐105 can also implement the viability and migration of non-small lung cancer cells by upregulating the expression of Mcl-1.13 To the best of our knowledge, our current study has inaugurally determined the role of miR-105 in OSCC progression.
Nevertheless, some limitations are present in this study. Firstly, the prognostic value of miR-105 in OSCC patients requires further validation using external (and broader) cohorts. Secondly, the biological function of miR-105 in the progression of OSCC cells in vivo needs to be investigated. Lastly, the underlying mechanism of how miR-105 exerts its biological function in OSCC needs to be examined in more details in future studies.
In summary, this study demonstrates that miR-105 is upregulated in OSCC tissues and plays a critical role in the proliferation and invasion of OSCC cells. Our findings indicate that miR-105 may serve as a prognostic biomarker as well as a potential therapeutic target for patients with OSCC. Future in vivo and clinical studies are warranted to corroborate our current results.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39(2):297–304. doi:10.1002/hed.24589
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
3. Thomson PJ. Perspectives on oral squamous cell carcinoma prevention-proliferation, position, progression and prediction. J Oral Pathol Med. 2018;47(9):803–807. doi:10.1111/jop.12733
4. Sarode GS, Sarode SC, Maniyar N, Anand R, Patil S. Oral cancer databases: a comprehensive review. J Oral Pathol Med. 2018;47(6):547–556. doi:10.1111/jop.12667
5. Belcher R, Hayes K, Fedewa S, Chen AY. Current treatment of head and neck squamous cell cancer. J Surg Oncol. 2014;110(5):551–574. doi:10.1002/jso.23724
6. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
7. Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. doi:10.1615/CritRevEukaryotGeneExpr.2017019494
8. Sun Z, Shi K, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi:10.1186/s12943-018-0897-7
9. Li J, Zhang Z, Chen F, et al. The diverse oncogenic and tumor suppressor roles of microRNA-105 in cancer. Front Oncol. 2019;9:518. doi:10.3389/fonc.2019.00518
10. Shen Z, Zhou R, Liu C, et al. MicroRNA-105 is involved in TNF-alpha-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213. doi:10.1038/s41419-017-0048-x
11. Gao R, Wang Z, Liu Q, Yang C. MicroRNA-105 plays an independent prognostic role in esophageal cancer and acts as an oncogene. Cancer Biomarkers. 2020;27(2):173–180. doi:10.3233/CBM-190736
12. Li HY, Liang JL, Kuo YL, et al. miR-105/93-3p promotes chemoresistance and circulating miR-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017;19(1):133. doi:10.1186/s13058-017-0918-2
13. Jin X, Yu Y, Zou Q, et al. MicroRNA-105 promotes epithelial-mesenchymal transition of nonsmall lung cancer cells through upregulating Mcl-1. J Cell Biochem. 2019;120(4):5880–5888. doi:10.1002/jcb.27873
14. Shen G, Rong X, Zhao J, et al. MicroRNA-105 suppresses cell proliferation and inhibits PI3K/AKT signaling in human hepatocellular carcinoma. Carcinogenesis. 2014;35(12):2748–2755. doi:10.1093/carcin/bgu208
15. Zhang J, Wu W, Xu S, et al. MicroRNA-105 inhibits human glioma cell malignancy by directly targeting SUZ12. Tumor Biol. 2017;39(6):1010428317705766.
16. Shang JC, Yu GZ, Ji ZW, Wang XQ, Xia L. MiR-105 inhibits gastric cancer cells metastasis, epithelial-mesenchymal transition by targeting SOX9. Eur Rev Med Pharmacol Sci. 2019;23(14):6160–6169.
17. Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL, Campbell M. miR-105 inhibits prostate tumour growth by suppressing CDK6 levels. PLoS One. 2013;8(8):e70515. doi:10.1371/journal.pone.0070515
18. Ries J, Baran C, Wehrhan F, et al. Prognostic significance of altered miRNA expression in whole blood of OSCC patients. Oncol Rep. 2017;37(6):3467–3474. doi:10.3892/or.2017.5639
19. Sun C, Li J. Expression of MiRNA-137 in oral squamous cell carcinoma and its clinical significance. J BUON. 2018;23(1):167–172.
20. Cristaldi M, Mauceri R, Di Fede O, Giuliana G, Campisi G, Panzarella V. Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: current status and perspectives. Front Physiol. 2019;10:1476. doi:10.3389/fphys.2019.01476
21. Ogawa H, Nakashiro KI, Tokuzen N, Kuribayashi N, Goda H, Uchida D. MicroRNA-361-3p is a potent therapeutic target for oral squamous cell carcinoma. Cancer Sci. 2020;111(5):1645–1651. doi:10.1111/cas.14359
22. He L, Ping F, Fan Z, et al. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed Pharmacother. 2020;121:109553. doi:10.1016/j.biopha.2019.109553
23. Chen R, Zhang Y, Zhang X. MiR-1254 functions as a tumor suppressor in oral squamous cell carcinoma by targeting CD36. Technol Cancer Res Treat. 2019;18:1533033819859447. doi:10.1177/1533033819859447
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.