Back to Journals » OncoTargets and Therapy » Volume 10
MicroRNAs as a novel class of diagnostic biomarkers for the detection of osteosarcoma: a meta-analysis
Authors Liu H , Li P, Chen L, Jian C, Li ZH , Yu AX
Received 14 June 2017
Accepted for publication 17 August 2017
Published 1 November 2017 Volume 2017:10 Pages 5229—5236
DOI https://doi.org/10.2147/OTT.S143974
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Hong Liu,1 Ping Li,2 Liang Chen,1 Chao Jian,1 Zonghuan Li,1 Aixi Yu1
1Department of Orthopedics, Zhongnan Hospital of Wuhan University, Wuhan, China; 2Department of Surgery, Experimental Surgery and Regenerative Medicine, Ludwig-Maximilians University, München, Germany
Abstract: MicroRNAs (miRNAs) have been considered as promising diagnostic biomarkers for many diseases, especially for cancers. Numerous studies have reported the value of miRNAs in the diagnosis of osteosarcoma (OS), but the results vary greatly across different studies. Therefore, we conducted this meta-analysis to assess the prospective diagnostic value of miRNAs in diagnosing OS. All relevant articles from prior to July 28, 2017 were selected from PubMed, EMBASE, Web of Science, Cochrane Library, Chinese National Knowledge Infrastructure, and Wan-fang databases. The Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) was performed to assess the quality of each article. A random-effects model was used to pool the sensitivity and specificity of the positive likelihood ratio (PLR), negative likelihood ratio (NLR) and, diagnostic odds ratio (DOR) together with the area under the curve (AUC) to evaluate diagnostic values. Seventeen studies comprising 2,214 OS patients and 1,534 healthy humans were included in our meta-analysis. The pooled estimations indicated that the miRNAs had a high accuracy for diagnosing OS, with a sensitivity of 0.82, specificity of 0.88, PLR of 10.96, NLR of 0.20, DOR of 54.55, and AUC of 0.93. Twenty-five miRNAs were differentially expressed in OS, including 17 upregulated and 8 downregulated. These miRNAs were correlated with survival time, tumor size, cell differentiation, tumor node metastasis staging, metastasis, tumor/cell invasion, pathological type, and response to radiotherapy and chemotherapy. Several different miRNAs are expressed in OS, and some of them might be potential biomarkers for the early diagnosis of OS.
Keywords: osteosarcoma, miRNAs, diagnosis, biomarker, meta-analysis
Introduction
Osteosarcomas (OS) originate from the mesenchyme tissue and are the most common malicious bone tumor. They occur primarily in the metaphysis of long bones and always affect adolescents and children.1–3 The incidence of OS is ~5 per million among Asian populations.4 Although some effective treatments, such as advanced surgical methods, adjuvant chemotherapy and radiotherapy, have significantly increased the survival rate of patients with OS, its 5-year survival rate is not high.1,3,5 The main reasons are highly malignancy, strong invasiveness, and early pulmonary metastasis.6,7 Early diagnosis and treatment of cancer are still two big challenges for clinicians and, therefore, it has become necessary to find an effective method for the initial diagnosis of OS.
It is well established that the standard OS therapy consists of early operation combined with neoadjuvant and adjuvant chemotherapy, but once metastasis occurs, this type of treatment does not work well. Currently, the diagnosis of osteosarcoma depends primarily on X-ray, localized symptoms, and pathology results, and it is difficult to detect early metastasis. Therefore, it is urgent to find innovative approaches for detecting OS in patients at the initial stage and for developing more efficient therapeutic methods to achieve a better prognosis.
miRNAs are a class of small non-coding RNA molecules (containing ~22 nucleotides) that play a critical role in gene silencing and post-transcriptional regulation of gene expression and occur broadly in plants, animals, and some viruses.8,9 Researchers have found that ~30% of genes in the human genome are regulated by miRNAs10 and that miRNAs participate in tumor development, metastasis, angiogenesis, and immune reactions.4,11,12 Thus, miRNAs play very important roles in homeostasis and tumorigenesis.13,14 Recently, numerous reports have shown that miRNAs are promising diagnostic and prognostic tools for cancers.15–17 The miRNAs have been confirmed to be promising diagnostic and prognostic markers in nasopharyngeal carcinoma18 and glioma19 and in chronic lymphocytic leukemia,20 acute lymphoblastic leukemia,21 breast cancer,22–24 gastric cancer,25,26 prostate cancer,27,28 pancreatic cancer,29 and colorectal cancer.30
The miRNAs in OS have also been widely tested. Cellular miRNAs can be identified rapidly and accurately from tissue and blood, especially serum and plasma,31 but the results are controversial. For example, Cai et al showed that miR-195 may be an indicative and predictive marker for OS,32 but other researchers stressed that miR-222 can be used to predict OS.33 Until now, no confirmed agreement has been reached. In this study, we identified and reviewed the most recent literature on miRNA and performed a meta-analysis to evaluate the diagnostic and prognostic value of miRNAs for OS.
Patients and methods
Search strategy and study selection
We conducted a literature search in the PubMed, EMBASE, Web of Science, Cochrane Library, Chinese National Knowledge Infrastructure, and Wan-fang databases to identify original articles analyzing the diagnostic value of miRNAs in OS. The last retrieval was performed before July 28, 2017. The medical subject headings (Mesh) and the entry terms employed in the literature search included 1) “osteosarcoma” or “osteosarcoma tumor”, 2) “microRNAs” or “miRNAs” or “primary microRNA” or “circulating microRNAs” or “circulating miRNA”, and 3) “diagnosis” or “sensitivity and specificity” or “receiver operating characteristics” or “ROC curve” or “ROC analyses” or “predictive value”. There were no restrictions on the language or publication date. The reference lists of review articles and selected papers were also skimmed to identify any additional eligible studies. Retrieved articles were independently screened and carefully evaluated by two investigators (LH and LZH). Two initial investigators (LP and CL) and another investigator (JC) participated in the data extraction. A fourth investigator (YAX) resolved any disagreements. We extracted data from the selected studies and input them into tables based on dissimilar characteristics of interest.
Selection criteria
This meta-analysis was carried out according to the guidelines of the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement. Studies that met the following criteria were included: 1) studies that evaluated the diagnostic value of miRNAs in OS patients; 2) studies from which meaningful data, such as sensitivity, specificity, and AUC values, were obtained; 3) studies that included specimens such as serum, plasma, or peripheral blood, and studies that used quantitative real-time polymerase chain reaction; 4) every individual study that involved both patients with OS and healthy control groups; and 5) articles that were written in English or Chinese. Accordingly, studies that met any of the following criteria were excluded: 1) duplicate of data already included in the analysis; 2) non-original paper, such as conference abstracts, letters, and reviews; 3) studies unrelated to OS; 4) animal experiments; or 5) studies that lacked complete data. For studies with overlapping populations or data, only the most complete report was reflected in the present meta-analysis. The selection process and inclusion and exclusion outcomes are shown in Figure 1.
  | Figure 1 Flow diagram of the selected eligible studies according to the inclusion and exclusion criteria. |
Data extraction
Two investigators (LP and JC) examined the full texts of eligible publications and extracted relevant data using a standardized table. From every included study, the following information was extracted: 1) study features, including the first author’s surname, publication year, and country; 2) participants’ general features, including ethnicity, type of case and control groups, and number of specimens; 3) data needed for the diagnostic meta-analysis, including studied miRNAs, specimen, sensibility, and specificity; and 4) information needed for quality assessment.
Quality assessment
In this meta-analysis, we comprehensively evaluated the quality of each included study based on the revised Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) checklist. The QUADAS-2 tool is composed of 4 key domains: patient selection, index test, reference standard, and flow and timing. These are supported by signaling questions to aid in judging.34 Table 1 presents a summary of all included studies.
Statistical analysis
The analytical software Meta-Disc version 1.4 and the statistical software Review Manager (RevMan 5.3) were utilized to analyze the diagnostic value and to calculate the 95% CI. The data were considered statistically significant when two-sided p<0.05. To indicate substantial heterogeneity inconsistency, I2 was computed. An I2 value >50% indicated substantial heterogeneity. Based on the heterogeneity values, we used the random-effect model throughout the statistical tests to compute the data. To investigate the possible sources of heterogeneity, meta-regression analyses were carried out. A subgroup analysis of the specimen type was performed for sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and, diagnostic odds ratio (DOR). The summary receiver operating characteristic (SROC) curve and the area under the curve (AUC) were also used to predict sensitivity and specificity. Additional data from validation studies that were not appropriate for pooling are listed in Table 2.
Results
Basic characteristics of included studies
As shown in Figure 1, a total of 1,167 published records related to the diagnostic value of miRNAs in OS were retrieved by a primary literature search (n=1,167) and hand search (n=0). Duplicate records (n=288) were removed. Then, 831 of the remaining 879 articles that were about animal experiments, that included other diseases, that did not include blood samples, and that were case reports or reviews were excluded, and full texts of the remaining 48 articles were evaluated for eligibility. The data were not complete in 31 of the 48 remaining articles. Therefore, this meta-analysis was conducted based on the remaining 17 articles.32,33,35–49 The main characteristics of the included articles are presented in Table 1.
The publication years of the 17 included articles were 2017 (n=2), 2016 (n=5), 2015 (n=6), 2014 (n=3), and 2012 (n=1). The dominant ethnicity of the study subjects was Asian. The expression levels of miRNAs of the plasma or serum samples were analyzed by quantitative real-time polymerase chain reaction.
Diagnostic accuracy of miRNAs in distinguishing OS from healthy controls
Table 2 presents the accuracy of the miRNAs in differentiating OS from healthy controls. A total of 17 studies covering 25 types of miRNAs and involving 3,748 participants (2,214 OS and 1,534 healthy controls) were included in the pooled analysis. The pooled sensitivity (Figure 2A) and specificity (Figure 2B) of the miRNAs for the diagnosis of OS were 0.82 (95% CI: 0.80–0.85) and 0.88 (95% CI: 0.86–0.90), respectively. The PLR, NLR, DOR, and AUC for the total miRNA levels in the 17 studies (Figures 3 and 4), with the corresponding 95% CIs, were 10.96 (95% CI: 4.67–25.76), 0.20 (95% CI: 0.15–0.28), 54.55 (95% CI: 27.38–108.67), and 0.93 (95% CI: 0.92–0.94), respectively.
In this meta-analysis, we conducted subgroup analyses of blood and plasma samples. The analyses results showed between-study heterogeneity. For a single miRNA assay, the pooled sensitivity, specificity, PLR, NLR, DOR, and AUC were 0.79 (95% CI: 0.77–0.82), 0.82 (95% CI: 0.78–0.85), 9.27 (95% CI: 3.02–28.46), 0.25 (95% CI: 0.19–0.32), 36.13 (95% CI: 14.71–88.71), and 0.90 (95% CI: 0.88–0.92), respectively. To detect whether plasma-based assays had a better diagnostic accuracy than serum-based assays, we determined the number of studies that used plasma/serum sample: of the 25 studies that assessed miRNAs, 11 used plasma samples and 14 used serum samples. The sensitivity of the studies that used plasma was 0.85 (95% CI: 0.82–0.87) and the specificity was 0.93 (95% CI: 0.91–0.95), with a pooled DOR of 86.05 (95% CI: 32.76–225.99) and AUC of 0.98 (95% CI: 0.96–0.99). Nevertheless, for serum-based studies, the sensitivity was 0.80 (95% CI: 0.77–0.82) and the specificity was 0.83 (95% CI: 0.79–0.88), with a pooled DOR of 36.78 (95% CI: 14.79–91.5) and AUC of 0.89 (95% CI: 0.87–0.91).
Clinical studies that supplied the ratio of miRNAs in OS patients
The results showed that different miRNAs were differentially expressed in patients with OS and healthy controls, with 17 upregulated miRNAs (including miR-222, miR-27a, and miR-221), 8 downregulated miRNAs (including miR-326, miR-223, and miR-15), and the same miRNA expressed in different tissues in distinctly different patterns. Our research shows that the expression of miR-195 and miR-199a-3p is much higher in patients with OS and that their expression in plasma and serum exhibits distinctive difference between OS and control groups. The detailed results are shown in Table 3.
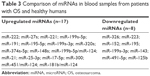  | Table 3 Comparison of miRNAs in blood samples from patients with OS and healthy humans |
Association between clinicopathological factors and differentially expressed miRNAs
The association between differentially expressed miRNAs and clinicopathological factors of OS patients is presented in Table 4. The individually expressed miRNAs were associated with survival time, tumor size, cell differentiation, tumor node metastasis staging, metastasis, tumor/cell invasion, pathological type, and response to radiotherapy and chemotherapy.
  | Table 4 Association between the expression of miRNAs and clinicopathological features of OS patients |
Discussion
Over the past several decades, we have achieved a lot in terms of the diagnostic methods for OS and significantly improved the treatment and prognosis of patients suffering from OS. However, the diagnosis of early stage OS and early lung metastasis is still challenging for clinicians. Finding other more sensitive and noninvasive methods for diagnosis is particularly important for the future treatment and diagnosis of OS.
The functional contributions of miRNAs in the development and progression of malignancies have resulted in the development of new diagnostic approaches. Many publications reported that miRNAs showed high diagnostic accuracy in distinguishing between patients with OS and healthy humans. Cao et al35 noted that miR-326 showed 83.7% sensitivity and 94.5% specificity. Dong et al36 reported the sensitivity and specificity for diagnostic accuracy of miR-223 to be 89.50 and 97.20%, respectively. Lian et al37 revealed that the miRNA panel (miR-195-5p, miR-199a-3p, miR-320a, and miR-374a-5p) could provide a high accuracy of OS in patients, with 91.10% sensitivity, 94.40% specificity, and an AUC of 96.08%. A reasonable diagnostic accuracy in discerning patients with OS and healthy humans is 83.7% sensitivity and 94.5% specificity. The AUC value is an effective indicator for the assessment system. An ideal test with perfect discrimination is at an AUC of 1.0. As the AUC value of a test gets closer to 1.0, the overall efficacy of the test will increase. Through the collection of relevant literature, we found that miRNAs could be used to screen OS patients compared to healthy controls with an AUC value of 0.90, which is very close to 1.0. This suggests that miRNAs have a relatively high ability to determine OS patients from healthy humans.
However, at this time, no agreement has been reached among several miRNA expression–profiling studies. Different protocols, measurement platforms, and small sample sizes could result in incomparable expression levels. Thus, we conducted this meta-analysis to systematically evaluate the diagnostic value of miRNAs for OS diagnosis. In our study, the results showed that there were 17 unregulated and 8 downregulated miRNAs in OS patients. Additionally, we found that miRNA assays using serum specimens, with a specificity of 0.83 and DOR of 36.78, greatly improved the diagnostic accuracy compared to plasma-based assays, with a specificity of 0.91 and DOR of 86.05. Although the results are inconsistent with previous research, it was indicated that sample types might play a critical role in investigating the utility of miRNAs as biomarkers in disease diagnosis. There were some specific miRNAs that were expressed differently in different studies. Liu et al40 reported that expression of miR-199a-3p was decreased in osteosarcoma patients compared to controls, whereas Lian et al37 reported that expression of the same miRNA was significantly higher in osteosarcoma patients than controls. The reason may be that the blood samples were taken at different times. Liu et al reported that fasting blood samples were obtained from patients prior to surgery and/or chemotherapy, whereas Lian et al37 reported that plasma samples were collected before surgery and 1 month after surgery. To confirm the best time for collecting the samples, future studies are needed. How the pathology, and response to radiotherapy and chemotherapy affect the expression of miRNAs and the specificity of these miRNA biomarkers should be further confirmed by comparing samples from patients with OS and patients with other types of cancers.
It was reported that miRNA expression profiles were considerably different between plasma and serum samples. Some researchers found that miRNA concentrations were higher in plasma.50 We also found that miRNAs assays using serum specimen, with a specificity of 0.83 and DOR of 36.78, greatly improved the diagnostic accuracy over plasma-based assays, with a specificity of 0.93 and DOR of 86.05. Based on these results, it is concluded that sample types may play a critical role in investigating the utility of circulating miRNAs as biomarkers in disease diagnosis.
Detecting the altered expressions of miRNAs could have several advantages: 1) we can get stable miRNAs from fresh tissues (cells and peripheral blood);51–53 2) each specific tissue is specifically expressed in miRNA; 3) miRNAs have been shown to be differentially expressed in patients with OS and normal humans; and 4) the expression of miRNAs combined with tumor stages has been determined.54,55 Thus, continued research on miRNAs could enable us to find innovative biomarkers and to better understand the molecular mechanisms of OS.
Our meta-analysis has multiple merits compared with previous studies. First, we included numerous miRNAs examined in preceding studies, which has increased the reliability of our assessment of their overall diagnostic value, compared with previous meta-analyses.56 Second, we conducted subgroup analyses to find potential sources of heterogeneity. Third, additional sensitivity analyses were utilized to confirm the strength of our results. However, in spite of these benefits, some limitations should be acknowledged. First, the literature sample size is small, which may decrease the level of evidence, so studies using larger sample sizes are needed to address the issue. Second, the biological functions of blood miRNAs are not very clear, which makes some data interpretation difficult. Third, although presented here as a potential diagnostic biomarker for OS, miRNAs possess numerous benefits and it is questionable whether these results can be implemented in clinical practice. Fourth, all of the data in this study are related to Asian countries, which is expected to result in heterogeneity. Finally, we could not obtain primary information regarding circulating miRNA expression data from these studies.
Conclusion
In conclusion, we found that many miRNAs play an important role in the early diagnosis of OS. Meanwhile, the miRNAs taken from plasma and serum samples could be promising noninvasive biomarkers for the early diagnosis of OS. Although the scheme seemed workable on paper, more experimental and clinical data need to be studied and collected step by step.
Disclosure
The authors report no conflicts of interest in this work.
References
Gorlick R, Janeway K, Lessnick S, Randall RL, Marina N; Committee COGBT. Children’s Oncology Group’s 2013 blueprint for research: bone tumors. Pediatr Blood Cancer. 2013;60(6):1009–1015. | ||
Rivera-Valentin RK, Zhu L, Hughes DP. Bone sarcomas in pediatrics: progress in our understanding of tumor biology and implications for therapy. Paediatr Drugs. 2015;17(4):257–271. | ||
Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. | ||
Bleyer A, Budd T, Montello M. Adolescents and young adults with cancer: the scope of the problem and criticality of clinical trials. Cancer. 2006;107(7 Suppl):1645–1655. | ||
Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11(12):849–864. | ||
Brennecke P, Arlt MJ, Campanile C, et al. CXCR4 antibody treatment suppresses metastatic spread to the lung of intratibial human osteosarcoma xenografts in mice. Clin Exp Metastasis. 2014;31(3):339–349. | ||
Bacci G, Rocca M, Salone M, et al. High grade osteosarcoma of the extremities with lung metastases at presentation: treatment with neoadjuvant chemotherapy and simultaneous resection of primary and metastatic lesions. J Surg Oncol. 2008;98(6):415–420. | ||
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. | ||
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. | ||
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. | ||
Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11(4):252–263. | ||
Hu H, Zhang Y, Cai XH, Huang JF, Cai L. Changes in microRNA expression in the MG-63 osteosarcoma cell line compared with osteoblasts. Oncol Lett. 2012;4(5):1037–1042. | ||
He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. | ||
Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. | ||
Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. | ||
Zeng X, Xiang J, Wu M, et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012;7(10):e46367. | ||
Wang Q, Li P, Li A, et al. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J Exp Clin Cancer Res. 2012;31:97. | ||
Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Si H, Sun X, Chen Y, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139(2):223–229. | ||
Schrauder MG, Strick R, Schulz-Wendtland R, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7(1):e29770. | ||
Ng EKO, Li R, Shin VY, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8(1):e53141. | ||
Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. | ||
Li C, Li JF, Cai Q, et al. MiRNA-199a-3p: a potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108(2):89–92. | ||
Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new biomarkers for prostate cancer. Br J Cancer. 2013;108(10):1925–1930. | ||
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. | ||
Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105(11):1733–1740. | ||
Menendez P, Padilla D, Villarejo P, et al. Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. 2013;108(6):369–373. | ||
Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67(3):875–884. | ||
Cai H, Zhao H, Tang J, Wu H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J Surg Res. 2015;194(2):505–510. | ||
Zhang Q, Wang S, Wang Y, Du Y. Serum level of microRNA-222 acts as a diagnostic and prognostic biomarker for osteosarcoma patients. Int J Clin Exp Pathol. 2016;9(4):4843–4848. | ||
Whiting PF, Rutjes AWS, Westwood ME, Mallett S, Deeks JJ, Reitsma JB. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. | ||
Cao L, Wang J, Wang PQ. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed Pharmacother. 2016;84:828–835. | ||
Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5(2):74–79. | ||
Lian F, Cui Y, Zhou C, Gao K, Wu L. Identification of a plasma four-microRNA panel as potential noninvasive biomarker for osteosarcoma. PLoS One. 2015;10(3):e0121499. | ||
Ma W, Zhang X, Chai J, Chen P, Ren P, Gong M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014;35(12):12467–12472. | ||
Wang NG, Wang DC, Tan BY, Wang F, Yuan ZN. Down-regulation of microRNA152 is associated with the diagnosis and prognosis of patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8(8):9314–9319. | ||
Ouyang L, Liu P, Yang S, Ye S, Xu W, Liu X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med Oncol. 2013;30(1):340. | ||
Ren H, Yang C, Su H, Li H. [Predictive effect of microRNA ratio in osteosarcoma]. J Int Oncol. 2014;41(9):708–711. Chinese. | ||
Tang J, Zhao H, Cai H, Wu H. Diagnostic and prognostic potentials of microRNA-27a in osteosarcoma. Biomed Pharmacother. 2015;71:222–226. | ||
Wang T, Ji F, Dai Z, Xie Y, Yuan D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer Biomark. 2015;15(5):543–550. | ||
Yang Z, Zhang Y, Zhang X, et al. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. | ||
Zhou G, Lu M, Chen J, et al. Identification of miR-199a-5p in serum as noninvasive biomarkers for detecting and monitoring osteosarcoma. Tumour Biol. 2015;36(11):8845–8852. | ||
Fujiwara T, Uotani K, Yoshida A, et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8(20):33375–33392. | ||
Xiao WF, Luo W, Xiao K, et al. [The expression of microRNA-125b in serum of patients with osteosarcoma and its inhibitory effect on proliferation]. Chin J Med Sci. 2016;8(8):49–52. Chinese. | ||
Wang Z, Jiang C, Chang X, Dai Y. Low miR-491-5p is an unfavorable prognostic marker for osteosarcoma. Int J Clin Exp Pathol. 2017;10(3):3304–3309. | ||
Liu JD, Xin Q, Tao CS, et al. Serum miR-300 as a diagnostic and prognostic biomarker in osteosarcoma. Oncol Lett. 2016;12(5):3912–3918. | ||
McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–840. | ||
De Carvalho IN, de Freitas RM, Vargas FR. Translating microRNAs into biomarkers: what is new for pediatric cancer? Med Oncol. 2016;33(5):49. | ||
Li H, Zhang K, Liu LH, et al. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol Lett. 2015;10(3):1662–1668. | ||
Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. | ||
Fan AC, Goldrick MM, Ho J, Liang Y, Bachireddy P, Felsher DW. A quantitative PCR method to detect blood microRNAs associated with tumorigenesis in transgenic mice. Mol Cancer. 2008;7:74. | ||
Zhao Y, Tu MJ, Yu YF, et al. Combination therapy with bioengineered miR-34a prodrug and doxorubicin synergistically suppresses osteosarcoma growth. Biochem Pharmacol. 2015;98(4):602–613. | ||
Wang X, Ning Y, Yang L, et al. Diagnostic value of circulating microRNAs for osteosarcoma in Asian populations: a meta-analysis. Clinical and experimental medicine. 2017;17(2):175–183. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





