Back to Journals » International Journal of General Medicine » Volume 16
MicroRNA Let-7i Regulates Innate TLR4 Pathways in Peripheral Blood Mononuclear Cells of Patients with Ankylosing Spondylitis
Authors Lu L, Fang H, Gu M, Wang H, Yu Q, Chen A, Gan KF
Received 18 November 2022
Accepted for publication 11 April 2023
Published 19 April 2023 Volume 2023:16 Pages 1393—1401
DOI https://doi.org/10.2147/IJGM.S397160
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Luca Testarelli
Liangjie Lu,1 Haiming Fang,1 Mengchao Gu,1 Huihan Wang,2 Qiuxia Yu,3 Aqiong Chen,3 Kai-feng Gan1
1Department of Orthopaedics, Li Huili Hospital Affiliated to Ningbo University, Ningbo, 315040, People’s Republic of China; 2Department of Orthopaedics, Zhengzhou Central Hospital Affiliated to Zhengzhou University, Zhengzhou, 450007, People’s Republic of China; 3Department of Rheumatology, Li Huili Hospital Affiliated to Ningbo University, Ningbo, 315040, People’s Republic of China
Correspondence: Kai-feng Gan, Tel +86-15724288924, Email [email protected]
Purpose: This study aimed to compare the changes in the expression of microRNA Let-7i in peripheral blood mononuclear cells (PBMCs) of patients with ankylosing spondylitis (AS) and the correlation between Let-7i and innate pro-inflammatory factors. It is necessary to search for a new biomarker to guide the prognosis of AS.
Methods: A total of 10 patients with AS and 10 healthy volunteers were selected as AS and control groups, respectively. The expression levels of Let-7i, Toll-like receptor 4 (TLR4), nuclear factor-κB (NF-κB), and interferon-gamma (IFN-γ) in PBMCs were detected by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting (WB) to explore the relationship between Let-7i and pro-inflammatory factors. Furthermore, the relationship between Let-7i and TLR4 was determined by the luciferase reporter technology.
Results: The expression level of Let-7i in PBMCs of patients with AS was significantly lower than that of healthy control. The expression levels of TLR4, NF-κB, and IFN-γ in PBMCs derived from patients with AS were significantly higher than those of healthy control. The results show that Let-7i manipulation can regulate lipopolysaccharide (LPS)-induced TLR4 and IFN-γ expression in CD4+ T cells of patients with AS. The overexpression of Let-7i in T cells of patients with AS can suppress TLR4 and IFN-γ LPS-induced expression levels of cellular mRNA and protein. Let-7i can directly interfere TLR4-3’untranslated region (UTR) sequence and regulate the expression of the TLR4 gene in Jurkat T cells.
Conclusion: Let-7i may be involved in the pathogenesis of AS, and Let-7i expression in PBMCs may be helpful for the diagnosis and treatment of AS in the future.
Keywords: ankylosing spondylitis, Let-7i, TLR4, innate inflammatory
Introduction
Ankylosing spondylitis (AS), as a prototype member of seronegative spondyloarthropathies, is an insidiously progressive and debilitating form of arthritis characterized by the involvement of the axial skeleton in the form of new bone formation and inflammation.1 Activated inflammatory responses can invade synovial joints, articular cartilage, tendons, ligaments, and attachment points of the ligament to cause fibrous and bony rigidity.2 The etiology and pathogenesis of AS are complex. Considering the long delay in diagnosis and insufficient available therapeutics, it is necessary to further understand the disease pathogenesis.3 Research on AS disease involves specific immune pathways, including antigen presentation, the modulator of nuclear factor-κB (NF-κB) activation, and T cell phenotype.4 The relevance of these immune-response pathways has been borne out in animal and human subject studies, particularly in response to novel diagnostic and therapeutic targets. The etiology of AS is thought to be a problem of autoimmune and/or auto-inflammation. Endogenous or infectious microbial triggers are critical to the onset and progression of AS disease regardless of the autoimmune or auto-inflammatory hypothesis model. Recent research has focused on the roles of microbiota and biomechanical stress in initiating and perpetuating inflammation.5
The mechanism of immunity interaction in establishing the observed state of cytokine dysregulation remains unclear. An in-depth understanding of the AS pathogenic process will optimize the application of current therapeutics and guide their future development. The biology of the implicated pathways needs to be studied by investigating AS patient subjects and model systems. More specifically, research should focus on the mechanism of inflammatory modulated pathways and the regulation of the responses to infectious and mechanical stressors in AS. The role of pathways in integrating mechanical stress, inflammation, and osteo-proliferation on AS needs further exploration.
How genetic variants predisposing to immune-mediated diseases relate to changes in immune system activity is an important question. The strong association of human leukocyte antigen (HLA)-B27 with this disease supports this hypothesis.6 Decades of effort have been made to identify other susceptibility genes. Recognition of an antagonistic antigen and proliferation of T cells to enhance immune responses are features of many immune-mediated diseases and contribute to promoting targeted inflammation.7 However, little evidence of such occurrence in patients with AS has been reported.
The innate immune system provides the first and immediate responses to foreign infectious and/or endogenous agents through pattern recognition receptors.8 TLR signaling associated with AS drives innate immune reactions and is a crucial pathway leading to AS disease.9 TLR ligands, particularly lipopolysaccharide (LPS), can activate NF-κB activity and promote inflammatory factor release, while NF-κB can affect the production of interferon,10 an NFκB activity calculator to delineate signaling crosstalk. Types I and II interferons can enhance NFκB via distinct mechanisms.11
MicroRNAs are endogenous non-coding single-stranded small-molecule RNAs with 18–25 nucleotides in length that play pivotal roles in multiple biological processes through post-transcriptional regulation and are closely related to human diseases.12 MicroRNAs can regulate immunologic functions and autoimmunity. Altered expression of microRNAs has been described under various pathological conditions, including autoimmune diseases.13 Emerging evidence suggests that miRNAs play a key role in regulating immunological functions, including innate and adaptive immune responses, the development and differentiation of immune cells, and the prevention of autoimmunity.14
Many studies have demonstrated that miRNA expression is altered in peripheral blood mononuclear cells (PBMCs) of patients with autoimmune diseases. Some microRNAs, including let-7i, miR-16, and miR-221,15 play essential roles in rheumatic diseases.16,17 Some microRNAs are expressed highly in patients with AS, ie, miR-29a, miR-335-5p, miR-27a, and let-7i.18 Exosomal let-7i has been reported to regulate T-cell-mediated autoimmune responses, while let-7i can suppress the induction of Treg cells by modulating their metabolism and function.
The study aimed to elucidate the association between let-7i expression, the pathogenic process of AS, and associated signaling pathways, to identify potential therapeutic targets, and to explore the potential roles of let-7i in AS.
Materials and Methods
Patients and Controls
At recruitment, demographic and clinical data, including age, gender, current medications, and medical history, were obtained. All patients were diagnosed with AS for the first time and received no treatment. Additionally, no subject had a clinically apparent infection or other inflammatory processes of any cause at recruitment. We excluded anyone who had used glucocorticoids (any dose) within the last 4 weeks. Ten patients with AS were recruited for this study. Ten age- and sex-matched healthy volunteers served as controls. This study was approved by the Ethics Committee of Li Huili Hospital Affiliated to Ningbo University (No. KY2020PJ002) and followed the Declaration of Helsinki. Each participant signed informed consent. Blood samples of patients with AS and healthy volunteers were collected, while patients with AS were graded by the Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) and the Bath Ankylosing Spondylitis Functional Index (BASFI).19
Isolation of RNA from T Cells
Heparinized venous blood of patients with AS and healthy volunteers was mixed with one-fourth volume of 2% dextran solution and incubated at room temperature for 30 min. Leucocyte-enriched supernatant was collected and layered over a Ficoll-Hypaque density gradient solution (specific gravity 1.077). After centrifugation at 250 g for 25 min, PBMCs were aspirated from the interface. Then, T cells were purified further by anti-human CD3 magnetic beads using IMag Cell Separation System (BD Bioscience, Franklin Lakes, NJ, USA). The T cell concentration was adjusted to 1×106/mL in RPMI-1640 containing 10% heat-inactivated fetal bovine serum (FBS), 2 mmol/l L-glutamine, penicillin (100 U/mL), and streptomycin (100 mg/mL) (10% FBS-RPMI) for further analysis. Total RNA, including miRNA from the T cells, was extracted using the mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. The RNA concentration was quantified using a NanoDrop Spectrophotometer.
Reverse Transcription (RT) of miRNAs
We converted all miRNAs into corresponding cDNAs in a one-step RT reaction. Briefly, a 10 mL reaction mixture containing miRNA-specific stem-loop RT primers (final 2 nM each), 500 mM deoxyribonucleotide (dNTP), 0.5 mL Superscript III (Invitrogen, Carlsbad, CA, USA), and 1 mg total RNA were used for RT reactions. The pulsed RT reaction was performed in the following conditions: 16°C for 30 min, followed by 50 cycles at 20°C for 30s, 42°C for 30s, and 50°C for 1s. After the RT reaction, the products were diluted 20-fold before further analysis.
Measurement of microRNA and mRNA Expression by qRT-PCR
The miRNA and mRNA expression levels were quantified using a qRT-PCR in this study. The miRNA RT kit (Applied Biosystems) was used to reverse RNA transcription into cDNA. One microliter of prepared RT product was used as a template for qPCR. Then, SYBR Master Mix, 200 nM forward primer, and 200 nM reverse primer were added for each qPCR. All reactions were duplicated on an ABI Prism 7500 Fast qRT-PCR system (Applied Biosystems). Conditions for the RT were 42°C for 30 min and 85°C for 15s, and the condition for qPCR was 95°C for 5 min, followed by 40 cycles of 95°C for 10s and 60°C for 35s. The expression of U6 small nuclear RNA (microRNA)/18S ribosomal RNA (mRNA) was used as an endogenous control for data normalization. The threshold cycle (Ct) refers to the cycle number at which the change of fluorescence intensity crosses the average background level of the fluorescence signal. The normalized microRNA/mRNA level was defined by the equation: 39 Ct after normalization by the expression of U6 small nuclear RNA/18S ribosomal RNA. The primers used for TLR-4 were 5’-ATTGGTGTGTCGGTCCTCAG-3’ (forward) and 5’-AGGCAGAGCTGAAATGGAGG-3’ (reverse). The primers used for IFN-γ were 5’-ACTAGGCAGCCAACCTAAGC-3’ (forward) and 5’-TTGGAAGCACCAGGCATGAA-3’ (reverse). The primers used for NF-κB were 5’-CAGCAGATGGCCCATACCTT-3’ (forward) and 5’-CACCATGTCCTTGGGTCCAG-3’ (reverse). The primers used for let-7i- were 5’-ACACTCCAGCTGGGTGAGGTAGTAGTTTGT-3’ (forward) and 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACAGCAC-3’ (reverse).
Western Blotting (WB) of Cell Lysates
T cells were lysed with 1% NP-40 (Sigma-Aldrich) in the presence of a proteinase inhibitor cocktail (Sigma-Aldrich). The cell lysates were electrophoresed and transferred to a polyvinylidene difluoride (PVDF) sheet (Sigma-Aldrich). After blocking, the membranes were incubated with primary antibodies followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Primary antibodies were against IFN-γ (1:500; cat. no. sc-373727), NF-κB (1:500; cat. no. sc-8414), and TLR-4 (1:500; cat. no. sc-52962) purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and β-actin (1:5000; cat. no. A5441) was purchased from Sigma-Aldrich as an internal control. As secondary antibodies, goat anti-rabbit, and goat anti-mouse immunoglobulin G (IgG) antibodies were obtained from Biovol Technologies (Biovol, Shanghai, China). The complexes formed were visualized after a chemiluminescence reaction (ECL; GE Healthcare, Little Chalfont, UK). The intensity of the respective band was semi-quantified by Image J.
Transfection of miRNA Mimics or Inhibitors into Jurkat Cells
Fresh isolated human T cells of patients with AS and healthy volunteers or Jurkat cells purchased from the American Type Culture Collection (Manassas, VA, USA) were infected with scrambled oligonucleotides, Let-7i mimics, or Let-7i inhibitors using recombinant Lenti-virus (Novobiotech, Shanghai, China). The working concentrations of Let-7i mimics and mimic-NC were 50 nM, and those of Let-7i inhibitors and inhibitor-NC were 100 nM. The expression of miRNA in miRNA-mimic or miRNA inhibitor was determined after culturing for 24 h at 37°C in a humidified atmosphere containing 5% CO2. These cells were then collected to analyze protein expression and mRNA transcription.
Functional Analysis of Let-7i in Human T Cells by Transfection with Let-7i Mimic or Inhibitor
T cells isolated from patients with AS were infected with Let-7i mimics/inhibitor recombinant Lenti-virus and then were cultured (i) in culture medium only, (ii) in 100 ng/mL LPS (Sigma-Aldrich) for 24 h at 37°C in a humidified atmosphere containing 5% CO2.20 Then, the immunocytes were collected to determine TLR4, NF-κB, and IFN-γ expression using qPCR and WB methods.
Bioinformatics and Luciferase Reporter Assay
Target genes for miRNAs were predicted using TargetScan web tools, as previously described.21 To certify the direct binding of let-7i to the 3ʹ-untranslated region (UTR), the wildtype (WT) or mutant reporter plasmid vector and up or up negative control were co-transfected in Jurkat cells using Lipofectamine™ 2000 (Invitrogen, Carlsbad, USA) in 24-well plates. After 48 h of transfection, cells were lysed. Subsequently, luciferase activity was measured using a Dual-Luciferase Reporter Assay Kit (Promega, USA) according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla activity and presented as relative luciferase activity. All assays were performed in triplicate.
Statistical Analysis
Statistical analysis was performed using SPSS software (V22.0) and GraphPad Prism 6. The statistical significance of differences between AS patients and control groups was analyzed by t-test (Clinical features, expression of microRNAs and mRNAs) and analysis of variance (ANOVA) (in vitro Let-7i manipulation experiments). When an ANOVA was conducted, the least significant difference (LSD) post hoc test was performed to show the individual differences. All data were represented as the mean standard deviation (SD), and a P-value less than 0.05 was considered statistically significant.
Results
Clinical Features of the Subject
A total of 20 samples (from 10 patients with AS and 10 healthy controls) were analyzed. The general information, general demographic information, and baseline clinical characteristics, including erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) score, Bath Ankylosing Spondylitis Functional Index (BASFI) score, disease duration, and morning stiffness of AS patient and control groups, are listed in Table 1. In this study, HLA-B27-positive patients with AS were collected, including six males and four females with an average age of 29.4±7.01 years. The healthy control group included six males and four females aged 30.3±5.91. There was no significant difference in gender and age between the two groups (P>0.05). The mean ± SD for ESR, CRP, BASDAI, BASFI, disease duration, and morning stiffness in patients with AS were 27.6±16.2 mm/h, 18.46±9.5 mg/L, 4.66±0.68, 4.16±0.5, 6.6±2.1 years, 33±13.4 minutes, respectively.
 |
Table 1 Demographics and Clinical Data on Ankylosing Spondylitis (AS) Patients and Healthy Volunteers |
Expression of Let-7i, TLR4, NF-κB, and IFN-γ in PBMCs of Patients with AS
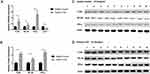
The AS is considered to be involved in inflammatory responses. Considering the diagnostic delay and insufficient therapy, a greater understanding of inflammation-related pathogenic progress of AS in patients’ blood is required. The expression of TLR4, NF-kb, and IFN-γ reflects the inflammation responses of immunocytes. The expression of Let-7i, TLR4, NF-kb, and IFN-γ in the PBMCs of patients with AS was detected using RT-PCR. Compared with the control group, the expression level of Let-7i in patients with AS’ PBMCs decreased significantly, while inflammatory genes, including TLR4, NF-kb, and IFN-γ, were strongly overexpressed in patients with AS (Figure 1A). To confirm the mRNA results of TLR4, NF-kb, and IFN-γ, their protein levels in PBMCs were investigated. The expression levels of TLR4, NF-kb, and IFN-γ strongly increased in patients with AS (Figures 1B–D). The results were consistent with their mRNA expression.
Let-7i Manipulation Regulated LPS-Induced TLR4 and IFN-γ Expression in CD4+ T Cells of Patients with AS
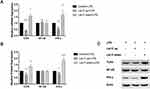
The microRNA Let-7i is negatively correlated with the above several inflammatory genes in patients with AS, suggesting that Let-7i may play an inflammatory regulatory role in AS. Let-7i overexpression/interference Lenti-virus was infected to CD4+ T cells of patients with AS to manipulate Let-7i levels. LPS induced the activation of TLR4, NF-kb, and IFN-γ of these immunocytes. The overexpression of Let-7i in T cells of patients with AS only suppressed TLR4 and IFN-γ LPS-induced expressive levels, while Let-7i interference promoted all three gene expressions induced by LPS (Figure 2A). To confirm qPT-PCR results, the protein levels of TLR4, NF-kb, and IFN-γ in LPS-treated CD4+ T cells of patients with AS were investigated. Only TLR4 and IFN-γ were significantly regulated by Let-7i (Figures 2B and C), consistent with their mRNA levels.
Let-7i Directly Interfered TLR4-3’UTR Sequence to Regulate the Expression of the TLR4 Gene in Jurkat T Cells
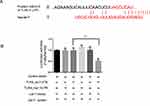
Let-7i is involved in the LPS-mediated TLR4 activation, indicating that Let-7i may directly interfere with the gene of the TLP4 pathway. The potential target sequence of TLR4 and IFN-γ was analyzed using the TargetScan web tool, and a 3’UTR sequence of TLR4 perfectly matched with Let-7i. Then, the Luciferase reporter of TLR4 3’UTR was designed to confirm the direct interference with Let-7i. With the co-transfection of Let-7i overexpression/interference Lenti-viruses into Jurkat T cells, WT/Mutant TLR4 3’UTR reporter indicated different self-luminescence results. Let-7i overexpression inhibited the luminescence from the WT reporter without affecting the Mutant reporter (Figure 3). The results showed that Let-7i directly affected the TLR4 expression by interfering TLR4 3’UTR region.
Discussion
In this study, we detected the expression levels of microRNA Let-7i and crucial regulators in the immune system, ie, TLR4, NF-κB, and IFN-γ, in PBMCs of patients with AS. The results revealed that Let-7i was down-regulated, and TLR4, NF-κB, and IFN-γ were up-regulated in PBMCs of patients with AS compared with healthy controls. It is concluded that Let-7i can regulate the expression of TLR4 and IFN-γ in CD4+ T cells of patients with AS.
TLR4, NF-κB, and IFN-γ are common inflammatory genes with biological roles in anti-infection, anti-tumor, and immune regulation. Under disease conditions, increased inflammatory factors can further cause tissue damage and exacerbations. As an autoimmune inflammatory disease, AS is closely related to inflammatory factors. It has been reported that TLR4, NF-κB, and IFN-γ are involved in the pathogenesis of AS.22
As TLR-4 is the prime cellular pattern recognition sensor for microbial pathogens, LPS can induce TLR-4 activation to produce pro-inflammatory cytokines in innate immune systems.23 Increased TLR-4 protein expression in PBMCs of patients with AS has been detected by flow cytometry and qRT-PCR in previous studies.24 The result is consistent with our results.
IFN-γ is a key proinflammatory cytokine that has been shown to be elevated in the serum of patients with AS.25 In this study, the IFN-γ mRNA and protein expression levels were increased in AS PBMCs. Furthermore, a correlation between let-7i and the mRNA expression of IFN-γ in patients with AS was determined. This result is consistent with the finding that let-7i up-regulate IFN-γ production.
The role of microRNAs in autoimmune diseases is receiving increasing attention, and recent studies have shown the dysregulation of various microRNAs in AS.16 Our present results further support the hypothesis that misregulated regulatory T cells are involved in pathological changes in patients with AS through the aberrant expression of specific microRNAs. Decreased expression of let-7i enhanced both IFN-γ mRNA expression and production in CD4+ LPS-stimulated T cells compared with non-stimulated T cells of patients with AS. Furthermore, qPCR and WB results showed that both messenger RNA and protein expression levels of TLR-4 and IFN-γwere significantly changed in let-7i-affected cell transfection studies. The data show that LPS-induced TLR-4 and IFN-γ expression in AS T cells can be regulated by Let-7i. Our results may prove that the bacterial LPS can still promote IFN-γ production in abnormal T cells of AS up-regulated by Let-7i. Microbe infection may be involved in AS pathogenesis, activating TLRs to elicit inflammatory reactions and ectopic bone formation in AS spine. However, it is still premature to conclude that bacterial infection is the leading cause of AS.
In addition, another prominent finding in our study was that the let-7i on the 3’UTR site of TLR-4 could specifically up-regulate TLR-4 expression in T cells. However, one microRNA can repress the post-transcription of hundreds of target genes.26 Other microRNA molecules may also be involved in the T-cell signaling pathway targeted by let-7i. Therefore, the downstream molecular mechanism of decreased let-7i expression stimulating CD4+ regulatory T immune response needs to be further studied.
Genome-wide association studies of AS involve specific immune pathways, including control of NF-κB activation.4 Our results have shown that NF-κB was up-regulated in PBMCs, but non-regulated in CD4+ T cells of patients with AS. Increased cytokine production or implication of a cytokine receptor locus does not always show good therapeutic efficacy.27 Prior stratification of individual patients based on the underlying pathogenesis may optimize treatment.
The expression level of Let-7i in PBMCs of patients with AS was negatively correlated with the expression levels of TLR4, NF-κB, and IFN-γ. The results suggest that Let-7i down may play an essential role in the onset and development of AS disease. Decreased expression of let-7i in AS T cells participates in the immune pathogenesis of AS by enhancing the TLR4/IFN-γ inflammatory response. Therefore, Let-7i may provide a new direction for diagnosing and treating AS.
Funding
This study was supported by the Medical and Health Plan of Zhejiang(No.2023KY1025), “HuiLi Funded” Research Project(2022YB009) and Medical Science and Technology Project of Ningbo, China (No.2019Y04).
Disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. The authors report no conflicts of interest in this work.
References
1. Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis--insights into pathogenesis. Nat Rev Rheumatol. 2016;12(2):81–91. doi:10.1038/nrrheum.2015.133
2. Simone D, Al Mossawi MH, Bowness P. Progress in our understanding of the pathogenesis of ankylosing spondylitis. Rheumatology. 2018;57(suppl_6):vi4–vi9. doi:10.1093/rheumatology/key001
3. Bond D. Ankylosing spondylitis: diagnosis and management. Nurs Stand. 2013;28(16–18):52–9; quiz 60. doi:10.7748/ns2013.12.28.16.52.e7807
4. Smith JA. Update on ankylosing spondylitis: current concepts in pathogenesis. Curr Allergy Asthma Rep. 2015;15(1):489. doi:10.1007/s11882-014-0489-6
5. Babaie F, Hasankhani M, Mohammadi H, et al. The role of gut microbiota and IL-23/IL-17 pathway in ankylosing spondylitis immunopathogenesis: new insights and updates. Immunol Lett. 2018;196:52–62. doi:10.1016/j.imlet.2018.01.014
6. Jethwa H, Bowness P. The interleukin (IL)-23/IL-17 axis in ankylosing spondylitis: new advances and potentials for treatment. Clin Exp Immunol. 2016;183(1):30–36. doi:10.1111/cei.12670
7. Michonneau D, Latis E, Curis E, et al. Metabolomics analysis of human acute graft-versus-host disease reveals changes in host and microbiota-derived metabolites. Nat Commun. 2019;10(1):5695. doi:10.1038/s41467-019-13498-3
8. Koupenova M, Clancy L, Corkrey HA, Freedman JE. Circulating Platelets as Mediators of Immunity, Inflammation, and Thrombosis. Circ Res. 2018;122(2):337–351. doi:10.1161/circresaha.117.310795
9. Oliveira-Toré CF, Moraes AG, Martinez GF, et al. Genetic polymorphisms of toll-like receptors 2 and 9 as susceptibility factors for the development of ankylosing spondylitis and psoriatic arthritis. J Immunol Res. 2019;2019:1492092. doi:10.1155/2019/1492092
10. Aletaha S, Haddad L, Roozbehkia M, et al. M2000 (β-D-Mannuronic Acid) as a novel antagonist for blocking the TLR2 and TLR4 downstream signalling pathway. Scand J Immunol. 2017;85(2):122–129. doi:10.1111/sji.12519
11. Mitchell S, Mercado EL, Adelaja A, et al. An NFκB activity calculator to delineate signaling crosstalk: type I and II interferons enhance NFκB via distinct mechanisms. Front Immunol. 2019;10:1425. doi:10.3389/fimmu.2019.01425
12. Li M, He Y, Zhou Z, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47(phox)-oxidative stress pathway in neutrophils. Gut. 2017;66(4):705–715. doi:10.1136/gutjnl-2016-311861
13. Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity. 2016;44(5):955–972. doi:10.1016/j.immuni.2016.05.002
14. Pauley KM, Chan EK. MicroRNAs and their emerging roles in immunology. Ann N Y Acad Sci. 2008;1143:226–239. doi:10.1196/annals.1443.009
15. Reyes-Loyola P, Rodríguez-Henríquez P, Ballinas-Verdugo MA, et al. Plasma let-7i, miR-16, and miR-221 levels as candidate biomarkers for the assessment of ankylosing spondylitis in Mexican patients naïve to anti-TNF therapy. Clin Rheumatol. 2019;38(5):1367–1373. doi:10.1007/s10067-019-04509-1
16. Najm A, Blanchard F, Le Goff B. Micro-RNAs in inflammatory arthritis: from physiopathology to diagnosis, prognosis and therapeutic opportunities. Biochem Pharmacol. 2019;165:134–144. doi:10.1016/j.bcp.2019.02.031
17. Li Z, Wong SH, Shen J, Chan MTV, Wu WKK. The role of MicroRNAS in ankylosing spondylitis. Medicine. 2016;95(14):e3325. doi:10.1097/md.0000000000003325
18. Yang W, Yan X, Xia Q, et al. Predisposition of six well-characterized microRNAs to syndesmophytes among Chinese patients with ankylosing spondylitis. Mod Rheumatol. 2019;29(1):173–180. doi:10.1080/14397595.2018.1453277
19. Zochling J. Measures of symptoms and disease status in ankylosing spondylitis: Ankylosing Spondylitis Disease Activity Score (ASDAS), Ankylosing Spondylitis Quality of Life Scale (ASQoL), Bath Ankylosing Spondylitis Disease Activity Index (BASDAI), Bath Ankylosing Spondylitis Functional Index (BASFI), Bath Ankylosing Spondylitis Global Score (BAS-G), Bath Ankylosing Spondylitis Metrology Index (BASMI), Dougados Functional Index (DFI), and Health Assessment Questionnaire for the Spondylarthropathies (HAQ-S). Arthritis Care Res. 2011;63(Suppl 11):S47–58. doi:10.1002/acr.20575
20. Cui H, Xie N, Tan Z, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. 2014;44(7):2085–2095. doi:10.1002/eji.201344126
21. Coronnello C, Benos PV. ComiR: combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41(WebServer issue):W159–W164. doi:10.1093/nar/gkt379
22. Vanaki N, Aslani S, Jamshidi A, Mahmoudi M. Role of innate immune system in the pathogenesis of ankylosing spondylitis. Biomed Pharmacother. 2018;105:130–143. doi:10.1016/j.biopha.2018.05.097
23. Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8(5):388–403. doi:10.1038/cmi.2011.26
24. Kragstrup TW, Andersen T, Holm C, et al. Toll-like receptor 2 and 4 induced interleukin-19 dampens immune reactions and associates inversely with spondyloarthritis disease activity. Clin Exp Immunol. 2015;180(2):233–242. doi:10.1111/cei.12577
25. Wang H, Sun N, Li K, Tian J, Li J. Assay of peripheral regulatory Vδ1 T cells in ankylosing spondylitis and its significance. Med Sci Monit. 2016;22:3163–3168. doi:10.12659/msm.897126
26. Detassis S, Grasso M, Del Vescovo V, Denti MA. microRNAs make the call in cancer personalized medicine. Front Cell Dev Biol. 2017;5:86. doi:10.3389/fcell.2017.00086
27. Janský L, Reymanová P, Kopecký J. Dynamics of cytokine production in human peripheral blood mononuclear cells stimulated by LPS or infected by Borrelia. Physiol Res. 2003;52(6):593–598.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.