Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNA-708 Suppresses Cell Proliferation and Enhances Chemosensitivity of Cervical Cancer Cells to cDDP by Negatively Targeting Timeless
Authors Zou X , Zhu C , Zhang L , Zhang Y , Fu F, Chen Y, Zhou J
Received 13 August 2019
Accepted for publication 22 November 2019
Published 9 January 2020 Volume 2020:13 Pages 225—235
DOI https://doi.org/10.2147/OTT.S227015
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
Xinwei Zou,1–3,* Chenjie Zhu,1–3,* Lin Zhang,1 Yi Zhang,1 Fengqing Fu,2,3 Youguo Chen,1–3 Jinhua Zhou1–3
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China; 2Clinical Research Center of Obstetrics and Gynecology, Jiangsu Key Laboratory of Clinical Immunology, Soochow University, Suzhou, People’s Republic of China; 3Jiangsu Institute of Clinical Immunology, The First Affiliated Hospital of Soochow University, Suzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinhua Zhou; Youguo Chen
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Soochow University, 188 Shizi Road, Suzhou 215006, People’s Republic of China
Tel +86 512 6797 2022
Fax +86 21 6408 5875
Email [email protected]; [email protected]
Purpose: Cervical cancer is the fourth most common cause of cancer-associated mortality in women worldwide. Previous studies have reported that microRNAs (miRNAs) are involved in multiple biological aspects of cancer progression by regulating gene expression. Here, we investigated the role of microRNA-708 (miR-708) in cervical cancer.
Methods: The expression levels of miR-708 in cervical cancer tissues and paired-normal cervical tissues were tested by quantitative polymerase chain reaction (qPCR). The interaction between miR-708 and Timeless was identified by bioinformatics method, dual-luciferase reporter assay, and Western blotting. The effects of over-expression of miR-708 on cell proliferation and cisplatin sensitivity were determined by Cell Counting Kit-8 (CCK-8) and colony formation assay. Cell cycle and apoptosis were analyzed by flow cytometry. DNA damage induced by over-expression of miR-708 was determined by comet assay. Expression levels of the genes involved in repair of DNA damage were analyzed by Western blotting.
Results: MiR-708 was down-regulated in cervical cancer tissues compared with paired-normal cervical tissues. By bioinformatics method, Western blotting, and dual-luciferase reporter assay, we found that Timeless was a direct target of miR-708. Furthermore, miR-708 suppressed cellular viability, colony formation, promoted apoptosis, and induced DNA damage levels. MiR-708 also enhanced chemosensitivity of cervical cancer cells to cDDP via impairing the ATR/CHK1 signaling pathway.
Conclusion: We conclude that miR-708 suppresses cell proliferation, facilitates cisplatin efficacy, and impairs DNA repair pathway in cervical cancer cells. These results demonstrate that miR-708 might be a candidate therapeutic target for future cervical cancer therapy.
Keywords: cervical cancer, miR-708, Timeless, repair of DNA damage, chemotherapy
Introduction
Cervical cancer is one of the leading gynecological malignancies in women worldwide. In addition, more than 527,600 new cases and 265,700 deaths were attributed to cervical cancer worldwide in 2012.1 Although platinum-based chemotherapies are such major treatments for advanced cervical cancer, recurrence and distant metastasis have still remained the main causes of the low 5-year survival rate for patients with advanced cervical cancer.2 Therefore, it is of great importance to find out an effective molecular-targeted therapy for the improvement of prognosis.
Timeless gene is involved in circadian rhythmicity in Drosophila melanogaster and interacts with clock proteins.3 Additionally, an increasing number of studies have found that Timeless is over-expressed in several tumor types, including lung cancer, breast cancer, hepatocellular cancer, and cervical cancer.4,5–8 Furthermore, aberrant Timeless expression is associated with poor survival in lung cancer, breast cancer, and cervical carcinoma,4,6,8 indicating that Timeless plays a substantial role in these malignancies. Besides, recent studies have shown that Timeless is involved in the regulation of homologous recombination (HR) and none-homologous end joining (NHEJ) repair, which are two important methods to repair DNA damage by interacting with poly (ADP-ribose) polymerase-1 (PARP-1).9,10 Our previous study demonstrated that Timeless is aberrantly expressed in cervical cancer due to transcriptional activation by E2F1 and E2F4 (manuscript prepared), while the mechanism of Timeless over-expression has still remained elusive.
MicroRNAs (miRNAs), small non-coding RNAs of 20–22 nucleotides, recognize and bind with the 3ʹ-UTR of target mRNAs, leading to the cleavage and/or translational repression of mRNAs.11 It also has been demonstrated that miRNAs are involved in different cellular processes, including cellular differentiation, proliferation, oncogenesis, angiogenesis, invasion, and metastasis.11–14 Thus, the study of miRNAs may provide novel targets for anti-cancer therapies, regarding the correlation of expression of microRNA with multiple biological aspects of cancer progression.15–17 Diverse roles of miR-708 have been reported in different cancers, pro-tumorigenesis in bladder and colorectal cancers, while anti-tumorigenesis in Ewing’s sarcoma, glioblastoma multiforme, hepatocellular carcinoma, renal cell carcinoma, as well as breast, prostate, and ovarian cancers.18 However, the role of miR-708 in cervical cancer has still remained elusive.
In this study, we aimed to investigate the role of miR-708 in cervical cancer. We demonstrated that miR-708 was down-regulated in cervical cancer tissues compared with paired-normal cervical tissues. We also found that Timeless was a direct target of miR-708, and miR-708 could attenuate proliferation, promote apoptosis, and facilitate cisplatin sensitivity in cervical cancer cells. MiR-708 also impaired the ATR/Chk1 signaling pathway. Our findings might provide a valuable therapeutic target for cervical cancer.
Materials and Methods
Cell Lines and Primary Tissues
Human cervical cancer cell lines (SiHa, HeLa, C-33A, and Ca Ski) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were cultured either in Dulbecco’s modified Eagle's medium (DMEM) high glucose (HyClone Laboratories, Inc., Logan, UT, USA) or RPMI 1640 medium (HyClone Laboratories, Inc., Logan, UT, USA) supplied by 10% fetal bovine serum (FBS; Biosera, Shanghai, China) and 1% penicillin/streptomycin (Beyotime Institute of Biotechnology, Shanghai, China). The cells were cultured in a humidified incubator with 5% (v/v) CO2 at 37°C.
The primary-paired tissues were obtained from cervical cancer patients who admitted to the First Affiliated Hospital of Soochow University (Suzhou, China) after surgical resection. Paired-normal tissue was at least 2 cm away from any cancerous tissue and confirmed free of cancer or premalignant changes by two pathologists. The Ethics Committee of the First Affiliated Hospital of Soochow University approved this study. All patients signed written informed consent form prior to beginning the study.
RNA Extraction and Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was extracted from frozen tissues (cancer and paired-normal tissues) or cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For analysis of mRNA expression, RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for cDNA synthesis. Expression levels of genes were normalized to β-actin gene as an endogenous control. The sequences of qPCR primers were as follows: Timeless forward: 5ʹ-GTTTTGGCAATCTGCCTAAGGA-3ʹ, reverse: 5ʹ-GCAGCTCATACAAGGTTTCACT-3ʹ. β-Actin forward: 5ʹ-TGACGTGGACATCCGCAAAG-3ʹ, reverse: 5ʹ-CTGGAAGGTGGACAGCGAGG-3ʹ. For analysis of miRNA expression, Bulge-loopTM miRNA qPCR Primer Set (one RT primer and a pair of qPCR primers for each set) for miR-708 was designed by RiboBio Co., Ltd. (Guangzhou, China). The U6 gene was used as an endogenous control. Moreover, qPCR was carried out using Power SYBR Green PCR Master Mix (Takara, Kusatsu, Japan) according to the manufacturer’s instructions. Data analyses were performed using the 2−ΔΔCt method.
miRNA Transfection
Transfection of miRNA was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The miRNA mimics for cell transfection were purchased from GenePharma (Shanghai, China), and their sequences were as follows: Non-targeting siRNA (miR NC): 5ʹ-UGGUUUACAUGUCGACUAATT-3ʹ. MiR-708: 5ʹ-AAGGAGCUUACAAUCUAGCUGGG-3ʹ.
Bioinformatics Website and Luciferase Assay
Potential miRNAs targeting Timeless were analyzed by several bioinformatics methods: TargetScan (www.targetscan.org), DIANA Tools (http://diana.imis.athena-innovation.gr), miRanda database (www.microrna.org). Then, we calculate the intersections of these three lists by venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/).
The 3ʹ-UTR regions of timeless gene containing either wild or mutant predicted binding sites for miR-708 (88-93/276-281 GCUCCU mutated to CGAGGA) were cloned into psiCHECK-2 vectors. Cells were seeded in 24-well plates 1 day before transfection and then were co-transfected with 500 ng of the vectors and 100 nM of miR-708 mimics or mimic control duplexes using Lipofectamine 3000 reagent. After 24 hrs, luciferase activity assay was performed using the Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA).
Western Blotting
Total protein was extracted from whole cells with sodium dodecyl sulfate (SDS) buffer and 15 µg of protein lysates were separated by SDS-polyacrylamide gel (SDS-PAGE). Timeless (Abcam, Cambridge, UK), RAD51, P21, cleaved-caspase 3, γ-H2AX, CHK2, ATM, phosphor-ATM, phosphor-CHK2, phosphor-CHK1 (Cell Signaling Technology, Danvers, MA, USA), phosphor-ATR (Gentex Corp., Zeeland, MI, USA), ATR, CHK1 (R&D Systems, Minneapolis, MN, USA), and β-actin (Sigma-Aldrich, St. Louis, MO, USA) were incubated, respectively, at recommended concentration overnight at 4°C. Finally, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H+L) and goat anti-rabbit IgG (H+L) (Multi Sciences, Hangzhou, China) secondary antibodies were incubated for 1 hr at room temperature. The immunogenic proteins were visualized using an enhanced chemiluminescence (ECL) system; β-actin was used as a loading control.
Cell Viability and Cisplatin Sensitivity Assay
Cells (4000/well) were plated in 96-well plates and transfected with miRNA mimics after 24 hrs. At the time of 24, 48, 72, 96, and 120 hrs after transfection, the medium was replaced with Cell Counting Kit-8 (CCK-8; Doujindo Laboratories, Tokyo, Japan) and incubated at 37°C for 60 mins. Afterwards, absorbance at 450 nm was measured by Multiskan GO microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
In cisplatin sensitivity assay, 48 hrs after transfection, cells were treated with increased concentration of cisplatin for 24 h. The next steps were the same as mentioned above. The analyses of IC50 value were performed by GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA).
Colony Formation Assay
Here, 48 hrs after transfection, cells were digested and plated on 30-mm dishes (500 cells/dish). Two weeks later, the colonies were stained by crystal violet (Beyotime Institute of Biotechnology, Shanghai, China) for 20 mins after fixing by 4% paraformaldehyde for 20 mins. The colonies were counted by ImageJ software and the same settings were kept for all colony-counting experiments.
Flow Cytometry Analysis
In the present research, 72 hrs after cell transfection, cells were digested using the trypsin free of ethylenediaminetetraacetic acid (EDTA), and collected and re-suspended at a concentration of 106 cells/mL after three times of washing with 1 mL ice-cold phosphate-buffered saline (PBS). According to the instructions provided for the Annexin V-PE Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA), 5 μL of PE-Annexin V, and 5 μL of 7-AAD were added to each 100 μL of diluted binding buffer. After the cells were incubated at room temperature for 15 mins, cell apoptosis was detected by flow cytometer FC500/FC500-MPL (Beckman Coulter, Brea, CA, USA).
For cell cycle analysis, cells were added into 1 mL pre-cooled 75% alcohol (−20°C), and fixed at temperature of 4°C overnight after cell transfection for 72 hrs or 120 hrs. Cells were re-suspended in 500 μL propidium iodide (PI)/RNase Staining Buffer (BD Biosciences, San Jose, CA, USA) after twice washing with cold PBS. Following 30 mins of incubation in the dark at room temperature, cells were analyzed by flow cytometer FC500/FC500-MPL (Beckman Coulter, Brea, CA, USA).
Comet Assay
An alkaline comet assay was undertaken using a COMET assay kit (Bio-Techne, Minneapolis, MN, USA) after 72 hrs of cell transfection. At least 50 cells were captured using a fluorescence microscope (Leica Microsystems, Wetzlar, Germany) in each sample. Comet images were analyzed by CASP software (http://casplab.com), and the olive tail moment (OTM) and tail moment (TM) were considered for the analysis of parameters.
Statistical Analysis
Each experiment was independently repeated three times or more. Expression of miR-708 and Timeless in cervical cancer tissues and their paired-normal cervical tissues were compared by paired t-test. The associations between miR-708 expression levels and clinicopathological parameters in cervical cancer patients were evaluated using Fisher’s exact tests. The functional effect between the target transfectants and the controls was compared by the Student’s t-test. A two-tailed P value of less than 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. SPSS software (version 22.0; SPSS Inc) was used for statistical analysis.
Results
miR-708 Is Down-Regulated in Human Cervical Cancer Tissues
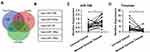
To explore the additional mechanisms of aberrant Timeless expression in cervical cancer, several bioinformatics methods (TargetScan, DIANA, miRanda) were applied to identify potential miRNAs targeting Timeless (Figure 1A). The results revealed that several miRNAs had potential bindings in the 3ʹ-UTR of Timeless (Figure 1B). Among these candidates, miR-708 was selected for further validation due to its potential roles in the repair of DNA damage and diverse roles in cancers.18 Additionally, qPCR was applied to identify the expression of miR-708 in 20 cervical cancer and paired-normal cervical tissues. A quantitative analysis revealed that miR-708 was significantly down-regulated in the cervical cancer tissues compared with the paired-normal cervical tissues (Figure 1C), whereas Timeless expression was upregulated in cervical cancer tissues (Figure 1D). The associations between miR-708 expression by qPCR and clinicopathological parameters in cervical cancer patients were shown in Table S1. The results guided us to presume the role of miR-708 in cervical cancer.
Timeless Is a Direct Target Gene of miR-708
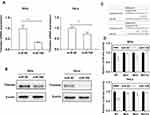
To detect the effects of miR-708 on the Timeless expression, miR-708 mimics were transfected in cervical cancer cells, and then Timeless mRNA and protein expression were detected after transfection. The results revealed that over-expression of miR-708 significantly inhibited the expression of Timeless compared with negative control miRNA (miRNA NC) in SiHa, HeLa, and Ca Ski cells (Figures 2A, B and S1).
The findings showed that the bases from 89 to 93 and 275 to 281 in the Timeless 3ʹ-UTR were perfectly complementary to the “seed” sequence of miR-708 (Figure 2C). To verify whether Timeless is a direct target of miR-708, a dual-luciferase reporter was constructed by inserting the wild-type 3ʹ-UTR of human Timeless gene into psiCHECK-2 vectors, and transfected into SiHa and HeLa cells together with either miR-708 mimics or miR NC. These results showed that the luciferase activity was significantly decreased by miR-708 compared to miR NC in both SiHa and HeLa cells after 24 hrs of post-transfection (Figure 2D). In order to clarify the binding sites for miR-708 in the 3ʹ-UTR of Timeless mRNA, we conducted equivalent luciferase constructs with single and double mutations in two putative binding sites. Compared with wild-type construct, double-mutation fully abolished the repression effect of miR-708, and mutation in site 1 or site 2, respectively, was able to partially resist against the repression effect of miR-708, while site 1 had a stronger resistance effect (Figure 2D). These results indicated that site 1 and site 2 both interact with miR-708, while site 1 has a stronger influence. Taken together, these results confirm that Timeless is a direct target of miR-708.
miR-708 Suppresses Proliferation of Cervical Cancer Cells and Promotes Apoptosis
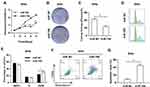
To investigate the biological function of miR-708 in cervical cancer, SiHa and HeLa cells were transfected either with miR-708 mimics or miR NC. As illustrated in Figures 3A and S2A, over-expression of miR-708 inhibited the proliferation of SiHa and HeLa cells compared with cells treated with miR NC. Consistently, the results of colony formation assay revealed that over-expression of miR-708 in both SiHa and HeLa cells significantly suppressed the colony formation (Figures 3B, C, S2B and C).
To further identify the potential role of miR-708 in cervical cancer, we analyzed cell cycle distribution after the over-expression of miR-708. Flow cytometry assay showed that cells were arrested at G2/M phase (Figures 3D, E, S2D and E). Apoptotic cells were also demonstrated by flow cytometry after miRNA transfection. The results revealed that the percentage of apoptotic cells was increased from 8.02% to 43.20% in SiHa, from 8.08% to 22.50% in HeLa, and from 16.70% to 39.20% in Ca Ski cells (Figures 3F, G, S2F and G). Furthermore, Western blotting detected increased cleavage of caspase-3, one of the key executioner of apoptosis,19 in SiHa and HeLa cells with over-expression of miR-708 compared with miR NC (Figure 4B). These data demonstrate that miR-708 suppresses cell proliferation and promotes apoptosis in cervical cancer.
miR-708 Induces DNA Damage and Impairs Activation of ATR Signaling Pathway
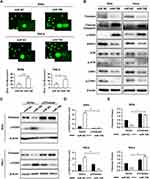
Since Timeless, the target of miR-708, was involved in repair of DNA damage, we investigated whether miR-708 also resulted in DNA damage. The comet assay was applied to measure DNA damage after miR-708 transfection. As displayed in Figures 4A, S3A, over-expression of miR-708 resulted in increased tail formation in SiHa, Ca Ski, and HeLa cells, indicating a higher level of DNA damage.
Next, Western blotting was applied to test the proteins involved in DNA damage and repair. The results suggested that over-expressed miR-708 led to increased level of phosphorylated histone H2AX (γH2AX) (Figure 4B). Additionally, we tested the expression of Rad51, as an important protein participating in homologous recombination of DNA during double-strand break repair.13 Rad51 expression was significantly decreased in SiHa and HeLa cells compared with the expression of miR NC (Figure 4B). Moreover, over-expression of miR-708 also attenuated the activation of ATR/CHK1 signaling pathway, as shown in Figure 4B. Previous studies have shown that miR-708 involved in repair of DNA damage by targeting Caspase-2, BMI1, and EYA3.20–22 We wondered whether miR-708 led to DNA damage by targeting Timeless. MiR-708 and pIRES2-EGFP-Timeless plasmid were co-transfected to operate a restore assay (Figures 4C, S3B). The expression of γH2AX induced by over-expression of miR-708 could be restored by simultaneous exogenous Timeless expression (Figures 4D, E, S3C and D), indicating that miR-708 can induce DNA damage at least partially by targeting Timeless.
miR-708 Enhances Chemosensitivity of Cervical Cancer Cells to cDDP
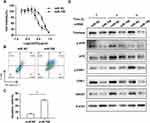
To determine whether miR-708 is involved in the regulation of cisplatin sensitivity, cells were treated with indicated concentration of cisplatin after transfected with miR-708 mimics or miR NC. Then, cell viability was measured and dose–response curves were plotted. The IC50 value was significantly decreased in SiHa cells with over-expression of miR-708 compared with negative control (5.126–18.26 μg/mL in miR NC vs 1.563–3.992 μg/mL in miR-708), indicating that miR-708 rendered SiHa cells more sensitively to cisplatin treatment (Figure 5A), which may provide a new strategy for sensitizing cervical cancer to cisplatin. As a result, cisplatin treatment resulted in more apoptotic cells in SiHa cells transfected with miR-708 mimics compared with control miRNAs, from 15.56% to 57.8% (Figure 5B and C). These results demonstrate that miR-708 potentiate cisplatin efficacy by inducing cell apoptosis.
We also attempted to investigate whether miR-708 regulates ATR/CHK1 signaling pathway in response to cisplatin. As shown in Figure 5D, the phosphorylated and total protein of ATR and CHK1 were both significantly reduced in cells with over-expressed miR-708 after cisplatin treatment, indicating an impaired ATR/CHK1 signaling pathway (Figure 5D). In addition, the expressions of Rad51 and P21 proteins were decreased in miR-708-overexpressed cells after cisplatin treatment (Figure 5D), which largely recapitulated the silencing of Timeless in cervical cancer (Paper in preparation).
Discussion
Previous studies have indicated that miR-708 is dysregulated in multiple cancer types. MiR-708 was shown to be up-regulated in bladder, colorectal cancer, and childhood B-cell acute lymphoblastic leukemia, while was down-regulated in breast, hepatocellular, prostate, ovarian, and renal cancer.22–29 However, the role of miR-708 in cervical cancer has still remained elusive. In the present study, we demonstrated that miR-708 was down-regulated in the cervical cancer tissues compared with paired-normal cervical tissues.
MiR-708 also plays a variety of roles in different cancers: pro-tumorigenesis in bladder and colorectal cancers, while anti-tumorigenesis in Ewing’s sarcoma, glioblastoma multiforme, hepatocellular carcinoma, renal cell carcinoma, breast, prostate, and ovarian cancers.18 Our phenotype analysis uncovered an anti-tumor role of miR-708 in cervical cancer. Over-expression of miR-708 attenuated the proliferation of cervical cancer cells. Besides, the higher number of Annexin V+/PE- cells as well as the increased level of cleaved-caspase 3 indicated the elevated level of apoptosis in miR-708-overexpressed cervical cancer cells.
It is of great importance to identify cancer-specific miRNAs and their target genes for understanding the role of miRNAs in tumorigenesis. In the present study, the Timeless gene sequence had two specific combination sites complementary to miR-708 sequence, as shown by bioinformatics analysis. We demonstrated that Timeless was indeed a direct target of miR-708 and site 1 as well as site 2, which both interacted with miR-708, while site 1 had a stronger influence, confirmed by luciferase reporter assays. Furthermore, expression of Timeless protein was significantly down-regulated by over-expression of miR-708 in cervical cancer cell lines. These findings suggest that miR-708 acts as a tumor suppressor in cervical cancer by directly targeting Timeless.
Previous studies have indicated that Timeless, recruited at replication fork, might play a significant role in DDR through interacting with CHEK1 and the ATR–ATRIP complex.30,31 Considering the fact that platinum-based chemotherapies induce DNA damage and replication stress,32,33 we thus explored whether miR-708 takes part in repairing process of DNA damage. As indicated by Western blotting and comet assays, over-expression of miR-708 increased the expression levels of phosphorylated histone H2AX (γH2AX) and OTM in cervical cancer cells, demonstrating a higher level of DNA damage. Besides, our results indicate that miR-708 plays role in the activation of ATR/CHK1 signaling pathway. Furthermore, over-expression of miR-708 decreased RAD51 expression, associating with tumor grade, metastases, treatment resistance, tumor relapse, and overall survival rate. The decrease of RAD51 expression may induce non-effective homologous recombination repair, resulting in mitotic catastrophe or apoptosis.34–36 Hence, we explored the role of miR-708 in cisplatin treatment and found that miR-708 potentiates cisplatin efficacy in cervical cancer cells.
Several limitations of our research should be expressed. Firstly, selective effect can avoid potential side effects and play a major role in targeted therapy, while we lack the data of normal cell lines, indicating whether over-expression of miR-708 has similar influences on normal cervical cells. For example, recent studies have revealed that inhibitors of ATR or CHEK1 could sensitize cancer cells to DNA damage, while normal cells are immune to them.37–39 Secondly, over-expression of miR-708 in conjunction with standard treatment may realize decreasing the dose of chemotherapy, thereby minimizing side effects. This hypothesis needs to be verified in our further experiments in cervical cancer xenograft models. Thirdly, the specific mechanism of how miR-708 acts in ATR/CHK1 signaling pathway and how RAD51 is regulated by miR-708 in cervical cancer require further investigation.
Conclusion
The present study demonstrated that miR-708 was down-regulated in the cervical cancer tissues. The overexpression of miR-708 in cervical cancer cells results in decreasing cell viability, as well as promoting apoptosis by directly targeting Timeless, a factor involved in DNA replication fork stability. Importantly, miR-708 could induce DNA damage and enhance the chemosensitivity of cervical cancer cells to cDDP through ATR/CHK1 signaling pathway. Identification of miR-708 targets may present new insights to the potential underlying mechanism in the treatment of cervical cancer. Collectively, our data demonstrated that miR-708 might be a candidate therapeutic target for future cervical cancer therapy.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81772773, 81672560, 81302275), Jiangsu Provincial Medical Youth Talent (Grant No. QNRC2016753), the Project of Jiangsu Provincial Maternal and Child Health Association (Grant No. FYX201709), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. SJCX18_0853), and the Jiangsu Provincial Maternal and Child Health Scientific Research Projeect (No. LGY2019043).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. doi:10.1016/S0140-6736(14)62038-9
2. Meijer CJ, Snijders PJ. Cervical cancer in 2013: screening comes of age and treatment progress continues. Nat Rev Clin Oncol. 2014;11(2):77–78. doi:10.1038/nrclinonc.2013.252
3. Ceriani MF, Darlington TK, Staknis D, et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285(5427):553–556. doi:10.1126/science.285.5427.553
4. Yoshida K, Sato M, Hase T, et al. TIMELESS is overexpressed in lung cancer and its expression correlates with poor patient survival. Cancer Sci. 2013;104(2):171–177. doi:10.1111/cas.2013.104.issue-2
5. Reszka E, Przybek M. Circadian genes in breast cancer. Adv Clin Chem. 2016;75:53–70.
6. Mao Y, Fu A, Leaderer D, Zheng T, Chen K, Zhu Y. Potential cancer-related role of circadian gene TIMELESS suggested by expression profiling and in vitro analyses. BMC Cancer. 2013;13:498. doi:10.1186/1471-2407-13-498
7. Elgohary N, Pellegrino R, Neumann O, et al. Protumorigenic role of timeless in hepatocellular carcinoma. Int J Oncol. 2015;46(2):597–606. doi:10.3892/ijo.2014.2751
8. Zhang W, He W, Shi Y, et al. Aberrant TIMELESS expression is associated with poor clinical survival and lymph node metastasis in early-stage cervical carcinoma. Int J Oncol. 2017;50(1):173–184. doi:10.3892/ijo.2016.3784
9. Young LM, Marzio A, Perez-Duran P, et al. TIMELESS forms a complex with parp1 distinct from its complex with TIPIN and plays a role in the DNA damage response. Cell Rep. 2015;13(3):451–459. doi:10.1016/j.celrep.2015.09.017
10. Xie S, Mortusewicz O, Ma HT, et al. Timeless interacts with PARP-1 to promote homologous recombination repair. Mol Cell. 2015;60(1):163–176. doi:10.1016/j.molcel.2015.07.031
11. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi:10.1155/2014/970607
12. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
13. McNutt M. Editorial expression of concern. Science. 2014;344(6191):1460. doi:10.1126/science.344.6191.1460-a
14. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
15. Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi:10.1073/pnas.0506654102
16. Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68(22):9125–9130. doi:10.1158/0008-5472.CAN-08-2629
17. Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi:10.1186/1476-4598-6-60
18. Monteleone NJ, Lutz CS. miR-708-5p: a microRNA with emerging roles in cancer. Oncotarget. 2017;8(41):71292–71316. doi:10.18632/oncotarget.v8i41
19. Kobayashi T, Masumoto J, Tada T, Nomiyama T, Hongo K, Nakayama J. Prognostic significance of the immunohistochemical staining of cleaved caspase-3, an activated form of caspase-3, in gliomas. Clin Cancer Res. 2007;13(13):3868–3874. doi:10.1158/1078-0432.CCR-06-2730
20. Song T, Zhang X, Zhang L, et al. miR-708 promotes the development of bladder carcinoma via direct repression of caspase-2. J Cancer Res Clin Oncol. 2013;139(7):1189–1198. doi:10.1007/s00432-013-1392-6
21. Robin TP, Smith A, McKinsey E, Reaves L, Jedlicka P, Ford HL. EWS/FLI1 regulates EYA3 in ewing sarcoma via modulation of miRNA-708, resulting in increased cell survival and chemoresistance. Mol Cancer Res. 2012;10(8):1098–1108. doi:10.1158/1541-7786.MCR-12-0086
22. Saini S, Yamamura S, Majid S, et al. MicroRNA-708 induces apoptosis and suppresses tumorigenicity in renal cancer cells. Cancer Res. 2011;71(19):6208–6219. doi:10.1158/0008-5472.CAN-11-0073
23. Song T, Xia W, Shao N, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11(4):905–911.
24. Piepoli A, Tavano F, Copetti M, et al. Mirna expression profiles identify drivers in colorectal and pancreatic cancers. PLoS One. 2012;7(3):e33663. doi:10.1371/journal.pone.0033663
25. Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Retracted: overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr Blood Cancer. 2013;60(12):2060–2067. doi:10.1002/pbc.v60.12
26. Ryu S, McDonnell K, Choi H, et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell. 2013;23(1):63–76. doi:10.1016/j.ccr.2012.11.019
27. Li G, Yang F, Xu H, Yue Z, Fang X, Liu J. MicroRNA-708 is downregulated in hepatocellular carcinoma and suppresses tumor invasion and migration. Biomed Pharmacother. 2015;73:154–159. doi:10.1016/j.biopha.2015.05.010
28. Saini S, Majid S, Shahryari V, et al. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer Res. 2012;72(14):3618–3630. doi:10.1158/0008-5472.CAN-12-0540
29. Lin KT, Yeh YM, Chuang CM, et al. Glucocorticoids mediate induction of microRNA-708 to suppress ovarian cancer metastasis through targeting Rap1B. Nat Commun. 2015;6:5917. doi:10.1038/ncomms6917
30. Engelen E, Janssens RC, Yagita K, Smits VA, van der Horst GT, Tamanini F. Mammalian TIMELESS is involved in period determination and DNA damage-dependent phase advancing of the circadian clock. PLoS One. 2013;8(2):e56623. doi:10.1371/journal.pone.0056623
31. Cho WH, Kang YH, An YY, Tappin I, Hurwitz J, Lee JK. Human tim-tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci U S A. 2013;110(7):2523–2527. doi:10.1073/pnas.1222494110
32. Rose PG, Bundy BN, Watkins EB, et al. Concurrent cisplatin-based radiotherapy and chemotherapy for locally advanced cervical cancer. N Engl J Med. 1999;340(15):1144–1153. doi:10.1056/NEJM199904153401502
33. Pearcey R, Miao Q, Kong W, Zhang-Salomons J, Mackillop WJ. Impact of adoption of chemoradiotherapy on the outcome of cervical cancer in Ontario: results of a population-based cohort study. J Clin Oncol. 2007;25(17):2383–2388. doi:10.1200/JCO.2006.09.1926
34. Wiegmans AP, Al-Ejeh F, Chee N, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5(10):3261–3272. doi:10.18632/oncotarget.v5i10
35. Short SC, Giampieri S, Worku M, et al. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro-Oncology. 2011;13(5):487–499. doi:10.1093/neuonc/nor010
36. Kiyohara E, Tamai K, Katayama I, Kaneda Y. The combination of chemotherapy with HVJ-E containing Rad51 siRNA elicited diverse anti-tumor effects and synergistically suppressed melanoma. Gene Ther. 2012;19(7):734–741. doi:10.1038/gt.2011.123
37. Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40(1):109–117. doi:10.1016/j.ctrv.2013.03.002
38. Lecona E, Fernandez-Capetillo O. Replication stress and cancer: it takes two to tango. Exp Cell Res. 2014;329(1):26–34. doi:10.1016/j.yexcr.2014.09.019
39. Merry C, Fu K, Wang J, Yeh IJ, Zhang Y. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2010;9(2):279–283. doi:10.4161/cc.9.2.10445
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.