Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNA-623 Inhibits Epithelial–Mesenchymal Transition to Attenuate Glioma Proliferation by Targeting TRIM44
Authors Cui D, Wang K, Liu Y, Gao J, Cui J
Received 19 February 2020
Accepted for publication 11 August 2020
Published 21 September 2020 Volume 2020:13 Pages 9291—9303
DOI https://doi.org/10.2147/OTT.S250497
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Dawei Cui,1 Kaijie Wang,2 Yan Liu,2 Junling Gao,3 Jianzhong Cui1,2
1Department of Surgery, Hebei Medical University, Shijiazhuang, Hebei 050017, People’s Republic of China; 2Department of Surgery, Tangshan Gongren Hospital, Tangshan, Hebei, 063000, People’s Republic of China; 3School of Basic Medical Science, North China University of Science and Technology, Tangshan, Hebei 063200, People’s Republic of China
Correspondence: Jianzhong Cui
Department of Surgery, Hebei Medical University, Shijiazhuang, Hebei 050017, People’s Republic of China
Email [email protected]
Objective: Glioma has the highest incidence among the different tumor types within the nervous system, accounting for about 40% of them. Malignant glioma has a high invasion and metastasis rate, which leads to the poor prognosis of patients. By targeting specific genes, microRNAs serve as key regulators in the epithelial–mesenchymal transformation (EMT) process, which could provide new insights into the treatment of glioblastomas (GBM). The detailed molecular role that miR-623 plays in GBM still remains unclear.
Materials and Methods: The level of miR-623 in GBM cells was evaluated by RT-PCR. The function of miR-623 overexpression on GBM cell proliferation, migration, and invasion was assessed by MTS, Transwell analysis, and colony formation assay. In addition, a mouse subcutaneous xenograft model was used to study in vivo effects. The binding between miR-623 and TRIM44 was verified by a dual-luciferase reporter assay and the regulatory function of miR-623 on EMT markers was evaluated using Western blot.
Results: The expression of miR-623 was repressed in the GBM cancer cell lines. MiR-623 overexpression or TRIM44 knockdown attenuated the proliferation, migration, and invasion of GBM cell lines. TRIM44 could facilitate the reverse suppression of EMT and miR-623 in GBM progression. MiR-623 was found to inhibit TRIM44 expression by directly binding to its 3ʹUTR. In addition, systemic delivery of miR-623 mimic reduced tumor growth and inhibited TRIM44 protein expression in tumor-bearing nude mice. Furthermore, our findings indicated that miR-623 overexpression or TRIM44 down-regulation impeded the proliferation and migratory ability of LN229 and U251MG glioma cells, and miR-623 attenuates TRIM44-induced EMT by directly targeting the 3ʹUTR of TRIM44, which could serve as preliminary research to identify potential therapeutic targets for future treatment of GBM.
Conclusion: Overall, microRNA-623 inhibits epithelial–mesenchymal transition to attenuate glioma proliferation by targeting TRIM44.
Keywords: GBM, hsa-miR-623, TRIM44, proliferation, EMT
Introduction
Glioma has been reported as the most prevalent primary craniocerebral tumor and originates from glial cells within the brain and spinal cord. At present, standard clinical treatment approaches for glioma are mainly based on surgical resection combined with chemotherapy and radiotherapy.1–3 New treatments have also appeared for gliomas: immunotherapy is mainly based on dendritic cells. The latest treatments include targeted therapy, the commonly used drugs are bevacizumab, gefitinib; CAR T cell therapy.4,5 Malignant glioma, with glioblastoma multiforme (GBM) being the most aggressive form, has high metastasis risk, which are the main reasons for the poor prognosis.6 Neovascularization in brain tumors correlates directly with their biological aggressiveness, degree of malignancy and clinical recurrence and inversely with the post-operative survival of patients.7,8 GBM has been reported to be the most angiogenic by displaying the highest degree of vascular proliferation and endothelial cell hyperplasia. Despite continuous advances in treatment methods and significant improvements in patient prognosis, breakthroughs in overall treatment strategies are still lacking.9–11 Therefore, finding a new treatment approach for glioma is a focus point of research.
Epithelial-mesenchymal transformation (EMT) refers to the transformation process of epithelial-derived tumor cells which were deprived of their polarity and acquire a mesenchymal phenotype6.12–14 EMT is an effective way for epithelial cells to acquire migration ability and plays an important role in invasion and metastasis in malignant tumors Studying EMT in glioblastomas has only started in recent years, and miRNAs have attracted much attention for regulating the invasion and progress of EMT. MiRNA is single-stranded non-coding RNA which has 21 to 23 bases in length and may participate in the cell proliferation, apoptosis invasion, migration, and cycle.14,15
Physiological processes, including plasma metabolism, cell differentiation, tumorigenesis and development, are closely related. Recently, miR-623 was identified as a tumor suppressor,16,17 whose expression was downregulated in some cancers such as lung gastric cells.18 However, the expression and regulatory role of miR-623 in GBM are yet to be clarified.
Tripartite pattern containing 44 (TRIM44) belongs to the protein family of TRIM which regulates a variety of pathological processes, such as neurodegenerative disorders, developmental abnormalities, or viral infections.19–21 Most E3 in them have ubiquitin ligase activity and can be post-translationally modified. Previous studies have found that the overexpression of TRIM44 in a variety of tumors could result in promoting cell proliferation, cell cycle progression and tumorigenesis. In addition, TRIM44 overexpression was also associated with the enhanced migratory and invasive capacities of tumor cells, thus facilitating tumor metastasis.22–24 Therefore, methods to inhibit TRIM44 expression is of great importance for controlling tumor development and metastasis.
Materials and Methods
Antibodies and Reagents
MiR-623 mimic and the respective controls were from GenePharma (Shanghai, China). The plasmid GV252-miR-623 expressing miR-623, lentiviral vector GV320-Ku80 and control vector were provided by GenePharma (Shanghai, China). The following siRNA target sequences were used to silence human siTRIM44 Sense: 5′-GAAUCAGUCGGAUACUCAUAG3′ and Antisense, 5′AUGAGUAUCCGACUGAUUCUG-3′. The oligonucleotides were chemically synthesized by GenePharma (Shanghai, China). Empty plasmid vector pENTER plasmid and TRIM44 overexpression plasmid pENTER-TRIM44 were purchased from Vigenebio Biosciences (Shandong, China). The oligonucleotides were chemically synthesized by GenePharma (Shanghai, China). Anti-TRIM44, anti-Vimentin, anti-MMP-2, anti-MMP-9, anti-β-actin, anti-E-cadherin and anti-N-cadherin were provided by Proteintech (Wuhan, China). Anti-Rabbit and anti-Mouse IgG were provided by Sigma (Shanghai, China).
Cell Culture
The human GBM cell line U87MG, LN229 and U251MG were purchased from the Cell Bank of the Chinese Academy of Sciences. Human normal brain glial cells (HEB) were acquired from the Cell Culture Center (Dongge Biotech, Beijing, China). The research ethics committee of Tangshan Gongren Hospital affiliated to Hebei Medical University approved the use of this cell. Cell line HEK 293T cells were obtained from ATCC (Manassas, VA). The medium used for cell culture was DMEM (GIBCO, USA) containing 10% fetal bovine serum (GIBCO, USA), and the incubation conditions included saturated humidity at 37°C and 5% CO2. When the cell coverage reached 80% to 90%, the culture passed and entered the logarithmic growth phase for subsequent experiments.
miRNA Transfection
Cells were transfected with miR-623 mimic and miRNA NC using Lipofectamine 3000 (Invitrogen, Carlsbad, USA). The procedures were followed according to the standard protocol set up by the manufacturer. After 48 h, the transfected cell samples were eligible for subsequent experiments.
Cell Proliferation Assays
The density of cell samples were approximately 3 x 103 cells/well. MST analysis was conducted at 0, 24, 48, 72 and 96 h after transfection. After 10 μL MST reagent was added to each well, the samples were incubated for 1 h at 37°C. For each sample, the optical density was read at 490 nm with a microplate reader (BioRad, USA).
Transwell Assay
Cell suspensions (8 × 104 cells/mL) of fetal bovine serum-free DMEM cultures with various transfection groups were harvested and inserted into the upper chamber of the Transwell chamber (BD Science, USA). 700 μL DMEM medium containing 20% fetal bovine serum was filled into the lower chamber. To assess cell invasion, the upper chamber was coated with Matrigel (BD Science) for 12 h. After normal incubation for 24 h, cells in the upper chambers were wiped and stained using crystal violet. Cells were counted in nine random fields.
Colony Formation Assay
At 1 × 103 cells per well, GBM samples in 6-well plates were transfected with miR-623 mimic, NC mimic. After 2 weeks, cells were processed with methanol fixation and Giemsa stain. The colony numbers were detected with a light microscope.
Quantitative RT PCR
Total RNA was acquired with miRNA extraction kit GenePharma (Shanghai, China). The purity and the amount of isolated RNA were determined with NanoDrop Spectrophotometer (Thermo Scientific, USA). Two μg total RNA was reversely transcribed into cDNA using BcaBest RNA PCR Kit (TaKaRa, Japan). Quantitative RT-PCR (qRT-PCR) was performed to determine gene expression using SYBR Green dye (Kangwei, China) on a BIO-RAD T100 real-time PCR system (BIO-RAD, USA). U6 miRNA and ACTIN were included as internal controls of miR-623 mRNA. The results were interpreted using the 2^-DDCt method.
Western Blotting Analysis
Whole-cell lysate was obtained after applying NP-40 containing a protease inhibitor cocktail and phosphatase inhibitor (Roche). An equal amount of lysate was separated by SDS-PAGE and then transferred to a PVDF membrane (Millipore, USA). Subsequent to blocking with 2% bovine serum albumin, the membrane was probed with anti-TRIM44 and anti-Actin (Proteintech, China), and then incubated with a second-generation culture medium combined with horseradish peroxidase. Goat anti-mouse IgG (1:2000) and goat anti-rabbit IgG (1:3000) were applied as secondary antibodies. The blots were visualized by an enhanced chemiluminescence solution (ECL; Pierce; Thermo Fisher Scientific).
Luciferase Reporter Assays
To verify the precise target of miRNAs, wildtype and mutant TRIM44 3ʹ-UTR were cloned into the pLUC vector, and then transfected into HEK293T cells together with miR-623 or NC mimic. Cell lysates were obtained at 48 h after transfection. Luciferase activity was measured using a dual-luciferase assay system (Promega, Madison, USA) according to the manufacturer’s instructions. Renilla-luciferase activity was assayed for normalization.
Acquire Stable-Transfected GBM Cell Lines
Lentiviral vector with luciferase and puromycin reporter containing overexpressed miR-623 and unrelated sequences were provided by GenePharma (Shanghai, China). HEK-293T cells were co-transfected with lentiviral plasmid and LipoFiter reagents to package recombinant lentivirus. At 72 h after transfection, lentiviral particles in cell culture supernatants were collected and filtered through a 0.45-μm filter (Millipore, USA). Recombinant lentivirus was processed with ultracentrifugation to concentrate it. To stabilize the transfected cell line, GBM cells were transduced with lentivirus (MOI=50) in the presence of 5 μg/mL polypropylene. After 24 h, the supernatant was discarded, and fresh DMEM medium was refilled into the sample. The infection efficiency was verified by RT-PCR at 96 h after transduction. A treatment of 2 μg/mL puromycin was applied for 2 weeks onto the cell samples for selection.
Tumorigenicity Assay in vivo
Male BALB/c athymic nude mice (6–7 weeks old) were offered by Beijing Huafukang Biotechnology (Beijing, China). All animal experiments and humane care were designed and supervised according to the Animal Experimentation Guidelines released by the Institute of Biomedicine. Nude mice received subcutaneous injection containing 5 × 106 transfected cells into the right hip. Tumor sizes were monitored every 4 days using calipers. Tumor volume was calculated as: V (cm3) = L (longest size) × W (shortest size)2/2. After 25 days, the mice were euthanized and tumor tissues were removed and weighed. A part of the tumor tissue was quick-frozen in liquid nitrogen and stored at −80°C for analysis of miR-623 and TRIM44 expression.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) software. Methods of statistical calculations included Student’s t-test, χ2 test, and analysis of variance (ANOVA). Data of all triplicate experiments are represented as mean ± SD. P <0.05 was considered as statistically significant.
Results
The Characteristics of miR-623 in GBM
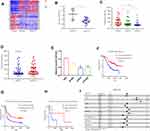
We first obtained 123 differentially expressed genes from CGGA database (grade IV, grade III compared with grade II, P < 0.05, and fold change >2 or <0.5) (Figure 1A). As shown in Figure 1C, compared with grade II patients, the expression of miR-623 was lower in grade III and grade IV patients. In addition, the expression of miR-623 in grade IV patients was lower than that in grade III patients. These revealed to us that miR-623 may be related to the grade of glioma and could also be used as a grade-related gene. And we downloaded the GEO dataset-GSE90603 and found that miR-623 was down-regulated in glioma tissues compared to normal tissues (Figure 1B). We got similar results in glioma cell lines. RT-RCR proved that the miR-623 levels in all three glioma cell lines (U87MG, LN229, U251MG) were lower compared to the normal glial cell line HEB (Figure 1E). We selected LN229 and U251MG cell lines for subsequent experiments. In view of the importance of IDH in gliomas, we also analyzed the relationship between miR-623 and IDH and found that the expression of miR-623 was higher in patients with IDH mutation (P = 0.0799). Followed, survival analysis showed that the expression of miR-623 was positively correlated to the survival of glioma patients. A higher expression of miR-623 was equal to a longer survival time (Figure 1F-H). Finally, we conducted a multifactor Cox survival analysis of miR-623, sex, age and other factors. The result is visualized in Figure 1I.
MiR-623 Inhibits GBM Cell Proliferation and Colony Formation
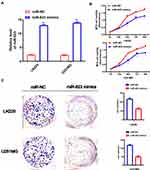
To study how miR-623 could interfere with the growth of GBM cells, we transfected miR-623 mimic into LN229 and U251 cells (Figure 2A). RT-PCR was conducted to verify successful miR-623 overexpression in GBM cells. We analyzed the proliferation of LN229 and U251MG cell lines with the transfection of miR-623 mimic and miR-NC by MTS assay. Validated by the inclusion of negative control, the cell proliferation was significantly inhibited by overexpressed miR-623 at 48 h,72 h and 96 h (Figure 2B). We followed-up with clone formation experiments with transfected glioma cell lines. Giemsa staining results showed that compared with the miR-NC group, overexpressed miR-623 significantly suppressed the colony formation in GBM cell lines (Figure 2C).
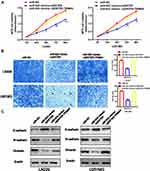
MiR-623 Inhibits GBM Cell Migration, Invasion and Affects the EMT Pathway
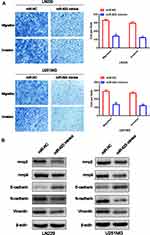
To investigate the regulatory function of miR-623 on the invasion and migration of GBM cells, we transfected miR-623 mimic into LN229 and U251 cells. Compared with the miR-NC control group, the number of migrated GBM cells was significantly reduced as evidenced by the Transwell experiment (Figure 3A). By performing Western blotting, we found that the expression of EMT-related markers, including MMP-2, MMP-9, N-cadherin and Vimentin, was significantly decreased in LN229 and U251 cells after the transfection of miR-623 mimic, while the expression of E-cadherin, which was an epithelial marker, was upregulated in the presence of overexpressed miR-623 (Figure 3B).
TRIM44 is ais a Direct Target of miR-623 in GBM
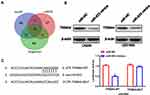
To probe into the mechanisms by which miR-623 modulates GBM cell functions, we selected potential downstream targets for miR-623 by various bioinformatics approaches, namely mirDIP, miRDB, and Targetscan, and found that TRIM44 could be targeted by miR-623 (Figure 4A and B). The bioinformatic results suggested that there was a putative miR-623 binding site located within the 3ʹ-UTR region of the TRIM44 transcript. Moreover, there was negative correlation between mir-623 and TRIM44(Supplementary Figure 1) To determine whether TRIM44 could interact with miR-623 via direct binding in human GBM cells, wildtype or mutant TRIM44 3ʹ-UTR plasmid was co-transfected with miR-623 mimic into 293T cells. At 48 h after the co-transfection, the luciferase activity was read. Overexpressed miR-623 resulted in a significant reduction of luciferase activity in the cells transfected with wildtype TRIM44-3ʹUTR. In contrast, the luciferase activity was not changed in the mutant TRIM44-3ʹUTR-transfected cells. In addition, qRT-PCR and Western blotting analysis further showed that miR-623 significantly reduced TRIM44 expression in terms of both mRNA and protein levels for 293T cells (Figure 4C).
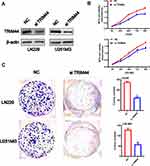
Knockdown of TRIM44 Can Inhibit Cell Proliferation and Colony Formation in GBM
To evaluate the function of TRIM44 on glioma cells, LN229 and U251MG cells were transfected with NC and si TRIM44, respectively. Western blot analysis confirmed that the protein level of TRIM44 was significantly reduced in the cell line transfected with si TRIM44 compared to cells transfected with NC (Figure 5A). MTS-way was used to detect the proliferation of glioma cells after knocking down TRIM44. It was demonstrated that cells transfected with si TRIM44 had a slower growth rate than NC transfected cells (Figure 5B). Moreover, clone formation experiments showed that TRIM44 knockdown can reduce the formation of glioma cell clones (Figure 5C).
Overexpression of TRIM44 Restores miR-623 Inhibited Cell Proliferation, Migration and EMT in GBM
To prove the functional relationship between TRIM44 and miR-623 in GBM cells, a rescue experiment was performed. First, the samples were randomly sorted into a miR-NC group, miR-623 overexpression plus empty plasmid vector group and miR-623 overexpression plus TRIM44 overexpression group, respectively. RT-RCR and Western blotting assay were conducted to evaluate the transfection efficiency. Subsequently, MTS and Transwell migration assays were performed (Figure 6A and B). The findings showed that overexpression of TRIM44 reversed the miR-623-mediated inhibition of proliferation and migration of LN229 and U251 cells. However, the protein level of E-cadherin was markedly down-regulated in miR-623 and TRIM44 co-overexpression GBM cell lines. Conversely, inhibited protein levels of N-cadherin and Vimentin were restored in TRIM44 overexpression GBM cell lines (Figure 6C).
TRIM44
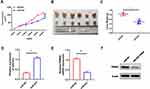
Finally, we investigated whether miR-623 could affect GBM growth in nude mice. U251MG cells were transduced with a lentiviral vector which stably miR-623 or a control lentiviral vector. After lentivirus infection, the down-regulation of miR-623 was examined with qRT-PCR. Then, we subcutaneously injected miR-623-silenced or control U251MG cells into BALB/c nude mice. From the 7th day after the injection onwards, the length and width of the tumors were measured every 4 days for up to 30 days. Overexpression of miR-623 significantly inhibited tumor growth in vivo (Figure 7A-C). Subsequently, we also measured the expression patterns for miR-623 and TRIM44 in tumor tissues. It was detected that miR-623 expression was up-regulated, and compared to the miR-NC group, TRIM44 expression in the miR-623 over-expression group was down-regulated at both mRNA and protein levels (Figure 7D-F).
Discussion
Accumulating evidence suggests that miRNAs exert key regulatory roles in the pathologic processes of many kinds of human tumors. By regulating the expression of their target genes, miRNAs can modulate various cellular biological behaviors such as cell growth, development, and proliferation. At the same time, miRNAs participate in tumor cell proliferation, invasion, migration, chemotherapy resistance, and immune escape by regulating gene transcription.25 In our study, we searched for differentially expressed genes in grade II, III, IV patients from CGGA. After that, we confirmed that miR-623 was lower expressed in glioma tissues compared to normal tissues. Moreover, the down-regulation of miR-623 was also related to tumor progression in GBM. These indicated that miR-623 may not only be used as a tumor biomarker, but a grade-related marker. The next study on the relationship between miR-623 and IDH also showed that there may be some connections between them, which needed to be explored in future. We also found that miR-623 was associated with survival time of GBM patients, whether overall survival, primary glioma survival or recurrent glioma survival. Our study showed the importance of miR-623 in GBM. Our current finding was consistent with the results in recent publications, which proved that the expression patterns of tumor-suppressive miRNAs were commonly decreased in GBM.
Epithelial–mesenchymal transition (EMT) is an effective way for epithelial cells to acquire migration ability, and it is an important process for more than 90% of epithelial cancer invasion and metastasis in malignant tumors.26,27 The fast-grow nature of tumor cells has become one of the typical characteristics of GBM, which may lead to the difficulties in cancer treatment and thus results in a poor prognosis. The molecular mechanisms regulating GBM cell growth and metastasis are still being explored. Therefore, finding key targets and understanding their functional roles in the development of GBM is the most important step for GBM treatment.28–30 Although many advances have been made to treat gliomas more effectively, the poor prognosis of the tumor has not yet been improved. The amount of research trying to find new therapeutic targets by uncovering the pathogenesis of glioblastoma so it can be treated more effectively is increasing.
Previous publications have shown that miR-623 could affect migration and invasion in different cancers. In our study, we analyzed the Chinese Glioma Genome Atlas (CGGA) database and found that miR-623 was associated with glioma survival. In general, the miR-623 expression was suppressed in glioma cell lines. After treating LN229 and U251MG glioma cells with miR-623 mimics, we performed MTS-way, invasion, migration, and clone formation tests. Previous studies have found that miR-623 mimics can attenuate the ability of gliomas to proliferate, invade, migrate, and form clones. It is worth noting that animal studies also have shown that tumor volume in the intracranial xenograft model is significantly reduced after miR-940 overexpression treatment, indicating its potential therapeutic value in glioma patients.31,32 Furthermore, there is accumulating evidence identifying a growing number of miRNAs that are involved in GBM metastasis. MiR-382 functions as a tumor suppressor against GBM development and metastasis and could be considered a potential drug source to improve the conventional GBM treatment strategies.33 These findings are in line with our results, which indicate that miR-623 functions as a tumor suppressor and could be a potential new target for GBM therapy.
Collectively, miRNAs are non-coding RNAs that play biological functions by binding to the 3ʹ-UTR of specific target genes. Through bioinformatic prediction, TRIM44 was pointed out as the downstream target gene for miR-623. Detection of the luciferase reporter gene showed that miR-623 inhibited wt TRIM44 3ʹ-UTR luciferase activity, but did not inhibit mt TRIM44 3ʹ-UTR. Previous studies have shown that abnormal expression of TRIM44 is also found in various human cancer types including lung cancer,34 melanoma,35 osteosarcoma,36 intrahepatic cholangiocarcinoma37 and renal cell carcinoma.38 Other studies also have found that TRIM44 is highly expressed in glioma cells and can be directly targeted by miR-101-3p and related to the EMT pathway of tumors.39 Our findings show that miR-623 mimics transfected glioma cells, and Western bolt showed that the levels of EMT-related markers, such as Vimentin, N-cadherin, MMP-2, MMP-9, were downregulated and E-cad expression increased. We suspect that the protective effect of miR-623 is based on the reduced EMT and enhanced E-cadherin level.
These results suggested that miR-623 might inhibit GBM growth in part by targeting TRIM44. However, this study also has limitations. Additional research is needed to further elucidate whether other factors also participate in the miR-623 signal axis and influence cancer progression.
Conclusions
We found that miR-623 inhibited the proliferation, migration, and invasion of GBM cells by activating TRIM44, as well as inhibiting tumor development in vivo. Our findings demonstrate that miR-623 has the potential to become a promising target for GBM treatment in the future and we are encouraging follow-up research.
Ethical Approval
The research ethics committee of Tangshan Gongren Hospital affiliated to Hebei Medical University approved the use of Human normal brain glial cells (HEB). All experiments involving mice have been approved Tangshan Workers Hospital Affiliated to Hebei Medical University Experimental Animal Ethics Committee and follow Institutional Animal Welfare Guidelines published in Chinese Academy of Medical Sciences.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Sampson JH, Gunn MD, Fecci PE, Ashley DM. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20(1):12–25.
2. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi:10.1007/s00401-005-0991-y
3. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi:10.1056/NEJMoa043330
4. Ratnam NM, Gilbert MR, Giles AJ. Immunotherapy in CNS cancers: the role of immune cell trafficking. Neuro-Oncology. 2019;21(1):37–46. doi:10.1093/neuonc/noy084
5. Bagley SJ, Desai AS, Linette GP, June CH, O’Rourke DM. CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro-Oncology. 2018;20(11):1429–1438. doi:10.1093/neuonc/noy032
6. Surawicz TS, Davis F, Freels S, Laws ER
7. Srivastava S, Jackson C, Kim T, Choi J, Lim MJC A. Characterization of dendritic cells and their role in immunotherapy in glioblastoma: from preclinical studies to clinical trials. 2019;11:4.
8. Finocchiaro G, Pellegatta S. Immunotherapy with dendritic cells loaded with glioblastoma stem cells: from preclinical to clinical studies. Cancer Immunol Immunother. 2016;65(1):101–109. doi:10.1007/s00262-015-1754-9
9. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi:10.3322/caac.20069
10. Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol. 2019;20(9):1100–1109. doi:10.1038/s41590-019-0433-y
11. PD-1 Blockade in GBM: uncovering response clues. Cancer Discov. 2019;9(6):687–688. doi:10.1158/2159-8290.CD-ND2019-001
12. Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi:10.1172/JCI38019
13. Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi:10.1038/ncb2976
14. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi:10.1038/nature03702
15. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
16. Ren F, Su H, Jiang H, Chen Y. Overexpression of miR-623 suppresses progression of hepatocellular carcinoma via regulating the PI3K/Akt signaling pathway by targeting XRCC5. J Cell Biochem. 2020;121(1):213–223. doi:10.1002/jcb.29117
17. Chen Y, Peng S, Cen H, et al. MicroRNA hsa-miR-623 directly suppresses MMP1 and attenuates IL-8-induced metastasis in pancreatic cancer. Int J Oncol. 2019;55(1):142–156. doi:10.3892/ijo.2019.4803
18. Jiang L, Yang W, Bian W, et al. MicroRNA-623 targets Cyclin D1 to inhibit cell proliferation and enhance the chemosensitivity of cells to 5-fluorouracil in gastric cancer. Oncol Res. 2018;27(1):19–27. doi:10.3727/096504018X15193469240508
19. Ong CA, Shannon NB, Ross-Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst. 2014;106:5. doi:10.1093/jnci/dju050
20. Li CG, Hu H, Yang XJ, Huang CQ, Yu XQ. TRIM44 promotes colorectal cancer proliferation, migration, and invasion through the Akt/mTOR signaling pathway. Onco Targets Ther. 2019;12:10693–10701. doi:10.2147/OTT.S228637
21. Chen Z, Lin TC, Bi X, et al. TRIM44 promotes quiescent multiple myeloma cell occupancy and survival in the osteoblastic niche via HIF-1α stabilization. Leukemia. 2019;33(2):469–486. doi:10.1038/s41375-018-0222-x
22. Zhou X, Yang Y, Ma P, et al. TRIM44 is indispensable for glioma cell proliferation and cell cycle progression through AKT/p21/p27 signaling pathway. J Neurooncol. 2019;145(2):211–222. doi:10.1007/s11060-019-03301-0
23. Xiong D, Jin C, Ye X, et al. TRIM44 promotes human esophageal cancer progression via the AKT/mTOR pathway. Cancer Sci. 2018;109(10):3080–3092. doi:10.1111/cas.13762
24. Zhou Z, Liu Y, Ma M, Chang L. Knockdown of TRIM44 inhibits the proliferation and invasion in papillary thyroid cancer cells through suppressing the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:98–103. doi:10.1016/j.biopha.2017.09.132
25. Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66(15):7390–7394. doi:10.1158/0008-5472.CAN-06-0800
26. Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development (Cambridge, England). 2012;139(19):3471–3486. doi:10.1242/dev.071209
27. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196.
28. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi:10.1038/nature03128
29. Wick W, Platten M. Understanding and treating glioblastoma. Neurol Clin. 2018;36(3):485–499. doi:10.1016/j.ncl.2018.04.006
30. Avril T, Vauleon E, Tanguy-Royer S, Mosser J, Quillien V. Mechanisms of immunomodulation in human glioblastoma. Immunotherapy. 2011;3(4 Suppl):42–44. doi:10.2217/imt.11.39
31. Luo H, Xu R, Chen B, et al. MicroRNA-940 inhibits glioma cells proliferation and cell cycle progression by targeting CKS1. Am J Transl Res. 2019;11(8):4851–4865.
32. Xu R, Zhou F, Yu T, et al. MicroRNA-940 inhibits epithelial-mesenchymal transition of glioma cells via targeting ZEB2. Am J Transl Res. 2019;11(12):7351–7363.
33. Wang J, Chen C, Yan X, Wang P. The role of miR-382-5p in glioma cell proliferation, migration and invasion. Onco Targets Ther. 2019;12:4993–5002. doi:10.2147/OTT.S196322
34. Luo Q, Lin H, Ye X, Huang J, Lu S, Xu L. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-κB signaling pathway. Int J Clin Oncol. 2015;20(3):508–517. doi:10.1007/s10147-014-0752-9
35. Wei CY, Wang L, Zhu MX, et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J Exp Clin Cancer Res. 2019;38(1):137. doi:10.1186/s13046-019-1138-7
36. Wang H, Fang ZL, Zhang GH, Ma X. TRIM44, a crucial target of miR-410, functions as a potential oncogene in osteosarcoma. Onco Targets Ther. 2018;11:3637–3647. doi:10.2147/OTT.S163163
37. Peng R, Zhang PF, Zhang C, et al. Elevated TRIM44 promotes intrahepatic cholangiocarcinoma progression by inducing cell EMT via MAPK signaling. Cancer Med. 2018;7(3):796–808. doi:10.1002/cam4.1313
38. Yamada Y, Kimura N, Takayama KI, et al. TRIM44 promotes cell proliferation and migration by inhibiting FRK in renal cell carcinoma. Cancer Sci. 2020;111(3):881–890. doi:10.1111/cas.14295
39. Leaf-nosed bat. Encyclopædia Britannica. Encyclopædia Britannica Online; 2009.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.