Back to Journals » OncoTargets and Therapy » Volume 12
microRNA-612 suppresses the malignant development of non-small-cell lung cancer by directly targeting bromodomain-containing protein 4
Authors Kang X, Kong F, Wu S, Liu Q, Yang C, Wu X, Zhang W
Received 3 February 2019
Accepted for publication 6 April 2019
Published 28 May 2019 Volume 2019:12 Pages 4167—4179
DOI https://doi.org/10.2147/OTT.S204004
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Gaetano Romano
This paper has been retracted.
Xiaowen Kang, 1,* Fanwu Kong, 2,* Shijie Wu, 3 Qiushuang Liu, 4 Chengcheng Yang, 1 Xiaomei Wu, 1 Wei Zhang 5
1Department of Respiration, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, People’s Republic of China; 2Department of Nephrology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, People’s Republic of China; 3Department of Respiration, General Hospital of Daqing Oil Field, Daqing, Heilongjiang 163000, People’s Republic of China; 4Department of Pharmacy, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, People’s Republic of China; 5Department of Respiration, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150000, People’s Republic of China
*These authors contributed equally to this work
Background: Aberrant expression of microRNAs (miRNAs) in non-small-cell lung cancer (NSCLC) has been reported. Dysregulation of miRNAs exerts tumor-suppressing or tumor-promoting actions on the pathology and biological behaviors of NSCLC. miR-612 is associated with many types of human cancer; however, the expression, potential roles, and regulatory mechanisms of miR-612 in NSCLC remain unclear.
Material and methods: Here, the expression level of miR-612 in NSCLC tissue specimens and a panel of cell lines were evaluated by RT-qPCR. Cell-Counting Kit 8, flow cytometry, Transwell migration and invasion, and in vivo tumor growth assays were performed to determine the functional role of miR-612 in malignant phenotypes of NSCLC cells. The molecular mechanism underlying the tumor-suppressive roles of miR-612 in NSCLC was investigated.
Results: miR-612 was expressed at low levels in NSCLC, and low miR-612 expression was significantly correlated with TNM stage and lymph node metastasis. NSCLC patients with low miR-612 expression had shorter overall survival rate than those with high levels. Exogenous miR-612 expression decreased proliferation, migration, and invasion, and promoted apoptosis of NSCLC cells in vitro. miR-612 upregulation hindered NSCLC tumor growth in vivo. Bromodomain-containing protein 4 (BRD4) was confirmed as a direct target gene of miR-612 in NSCLC cells. BRD4 was obviously overexpressed in human NSCLC tissues and inverse correlated with miR-612 expression. Inhibition of BRD4 expression simulated the tumor-suppressive functions of miR-612 overexpression in NSCLC cells. Reintroduction of miR-612 expression abrogated the miR-612-mediated suppressive effects on NSCLC cells. BRD4 upregulation inhibited activation of the PI3K/Akt pathway in NSCLC cells in vitro and in vivo.
Conclusion: This study supports the first evidence that miR-612 exerts tumor-suppressive roles in the aggressive behaviors of NSCLC cells in vitro and in vivo through direct targeting BRD4 and deactivating the PI3K/Akt pathway. Thus, miR-612 might be a promising target for anticancer therapies in patients with NSCLC.
Keywords: non-small-cell lung cancer, microRNA-612, bromodomain-containing protein 4, biomarker indicator
Introduction
Lung cancer is the third most common type of human malignancy and the leading cause of cancer-associated mortalities among both men and women globally.1 There are two main subtypes of lung cancer: non-small-cell lung cancer (NSCLC) and small cell lung cancer.2 NSCLC, which primarily includes squamous cell carcinoma, adenocarcinoma, and large cell carcinoma, is the most frequent type of lung cancer and accounts for approximately 80% of the total number of patients with lung cancer.3 Currently, the adoption of complex treatment, including surgical resection, radiotherapy, and targeted therapy, can remarkably improve the prognosis of patients with NSCLC; unfortunately, the long-term therapeutic outcomes remain unsatisfactory with a 5-year survival rate of <16.8%.4 Multiple risk factors have been demonstrated to be involved in the pathogenesis of NSCLC,5,6,7 including environmental pollution, smoking, and exposure to carcinogens; however, the exact mechanisms are not fully understood. Therefore, further investigation of the detailed mechanisms underlying NSCLC genesis and development is essential for developing effective therapeutic techniques and improving the prognosis of patients with NSCLC.
microRNAs (miRNAs) are a group of highly conserved, non-coding, and short RNAs, which are mainly expressed in plant, animal, and viral genomes.8 Over 2,000 mature miRNAs have been identified in the human genome, and they are predicted to be able to modulate the expression of one-third of all protein-coding genes.9 Mature miRNAs negatively regulate gene expression by sequence-specific binding to the 3′-UTRs of their target genes, ultimately resulting in mRNA degradation or translation suppression.10 miRNAs have broad effects on a wide range of biological behaviors, including cell proliferation, cell cycle, apoptosis, differentiation, metabolism, epithelial-mesenchymal transition, and metastasis.11,12,13 In particular, numerous miRNAs were identified to be dysregulated in NSCLC, including miR-374b,14 miR-500,15 miR-628,15 and miR-647.16 miRNAs may exert tumor-suppressive or oncogenic roles in the formation and progression of NSCLC based on the biological function of their target genes.17 This observation clearly indicates that miRNAs might be an effective therapeutic target for treating patients with NSCLC.
miR-612 has emerged as a cancer-associated miRNA in many types of human cancer, including bladder cancer,18 melanoma,19 colorectal cancer,20 ovarian cancer,21 and endometrial carcinoma.22 However, the expression and potential roles of miR-612 in NSCLC remain unclear, and the molecular mechanism responsible for the tumor-suppressive roles of miR-612 in NSCLC is still poorly understood. Hence, we evaluated miR-612 expression in NSCLC and examined its relationship with the clinical pathological features of patients with NSCLC. In addition, the biological effects of miR-612 on NSCLC cells and its underlying mechanism were explored. We demonstrated the important roles of miR-612 in the oncogenicity of NSCLC and that this miRNA might be a potential target for the therapy of patients with NSCLC.
Material and methods
Tissues samples
This study was approved by the Ethics Committee of The Second Affiliated Hospital of Harbin Medical University. Written informed consent was provided by all participants. Fifty-seven pairs of NSCLC tissues and corresponding adjacent normal tissues (ANTs) were collected from patients who underwent surgical resection in The Second Affiliated Hospital of Harbin Medical University. All tissue samples were quickly stored in liquid nitrogen after resection. None of the patients was treated with any additional treatment before surgery.
Cell culture and transfection
In total, five human NSCLC cell lines, including H522, H460, H1299, A549, and SK-MES-1, as well as a non-tumorigenic bronchial epithelium BEAS-2B were ordered from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). DMEM (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), penicillin (100 U/mL) and streptomycin (100 μg/mL) was utilized for the culture of all above cell lines. All cells were grown at 37°C in a humidified condition supplied with 5% CO2.
miR-612 agomiR (agomiR-612) and negative control agomiR (agomiR-NC) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). Small interfering RNA (siRNA) against the expression of bromodomain-containing protein 4 (BRD4) (BRD4 siRNA) and negative control siRNA (NC siRNA) were purchased from the Chinese Academy of Sciences (Changchun, China). BRD4 overexpression plasmid (pCMV-BRD4) and empty pCMV plasmid were prepared by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cells were plated into six-well plates at a density of 6 × 105 cells per well. The transfection experiments were mediated with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer’s protocol.
Reverse transcription-quantitative PCR (RT‑qPCR)
Total RNA from tissue samples or cultured cells was isolated using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. A NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to determine the concentration of total RNA. Total RNA was converted into first-strand complementary DNA (cDNA) using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression of miR-612 was quantified using a TaqMan MicroRNA qPCR assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). For the detection of BRD4 mRNA expression, an M-MLV (Promega, Madison, WI, USA) was adopted to synthesize cDNA, and the cDNA was then subjected to quantitative PCR using an SYBR Premix Ex Taq™ Kit (TaKaRa, Tokyo, Japan). The levels of miR-612 and BRD4 mRNA were normalized to U6 small nuclear RNA and GAPDH, respectively. Relative gene expression was calculated using the 2−ΔΔCt method.23
Cell-counting kit 8 (CCK-8) assay
The proliferation of NSCLC cells was examined using the CCK-8 assay. In detail, transfected cells were trypsinized 24 hrs post-transfection, counted, and inoculated into 96-well plates at an initial density of 3,000 cells per well. Cells were then incubated at 37°C for 0, 24, 48, and 72 hrs. The CCK-8 assay was carried out at every time point by adding 10 µL of CCK-8 reagent (Dojindo, Kumamoto, Japan) into each well. Following incubation at 37°C for 2 h, the absorbance at a wavelength of 450 nm was detected using a microplate reader (BioTek, Winooski, VT, USA).
Flow cytometry analysis
Flow cytometry analysis was utilized for the detection of cell apoptosis using an Annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit (Biolegend, San Diego, CA, USA). Forty-eight hours post-transfection, cells were harvested and washed thrice with ice-cold PBS (Gibco; Thermo Fisher Scientific, Inc.). Thereafter, cells were suspended in 100 µL of binding buffer that was supplemented with 5 µL of Annexin V-FITC and 5 µL of propidium iodide. After incubation for 30 mins at room temperature in the dark, the apoptosis rate was determined by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Transwell migration and invasion assays
Transfected cells were trypsinized and suspended into FBS-free DMEM medium. A total of 5×104 cells were added to the upper compartments of Matrigel-coated Transwell chambers (BD Biosciences) containing 8 μm pore-size polycarbonate membranes. The lower compartments of the chambers were filled with 600 μL of DMEM containing 20% FBS to serve as a chemoattract. Cells were then incubated at 5% CO2 at 37°C for 24 hrs. Subsequently, cells remaining in the upper chambers were removed with a cotton swab. The invasive cells that had invaded the lower side of polycarbonate membranes were fixed with 4% paraformaldehyde and stained with 0.05% crystal violet. The invaded cells were photographed under an inverted microscope (IX83; Olympus Corporation, Tokyo, Japan), and the invasion ability was expressed as the average number of invaded cells in five randomly selected microscopic fields per chamber. The Transwell migration assay was performed similarly to the invasion assay, except that the Transwell chambers were not coated with Matrigel.
In vivo tumor growth assay
H522 cells were transfected with agomiR-612 or agomiR-NC. After 24 hrs of culture, transfected cells were collected and injected subcutaneously into the flanks of BALB/c nude mice (Beijing Vital River Laboratory, Beijing, China). The volume of the xenograft was calculated using the following formula: length × width2×0.5. All nude mice were sacrificed 4 weeks post-inoculation. The xenograft was resected and weighed. All procedures involving animals were approved by the Experimental Animal Ethical Committee of The Second Affiliated Hospital of Harbin Medical University, and carried out in accordance with the Declaration of Helsinki and the guidelines of the Ethics Committee of The Second Affiliated Hospital of Harbin Medical University.
miR-612 target prediction
The putative targets of miR-612 were predicted using two publicly available databases: microRNA.org (
Luciferase reporter assay
The 3′-UTR sequences of BRD4 containing the predicted wild-type (Wt) or mutant (Mut) miR-612 binding sequences were amplified by Shanghai GenePharma Co., Ltd., and cloned into the pMIR-REPORT miRNA Expression Reporter vector (Ambion; Thermo Fisher Scientific, Inc.). The constructed luciferase reporter plasmids were defined as pMIR-Wt-BRD4-3ʹ-UTR and pMIR-Mut-BRD4-3ʹ-UTR, respectively. For the reporter assay, pMIR-Wt-BRD4-3ʹ-UTR or pMIR-Mut-BRD4-3ʹ-UTR, together with agomiR-612 or agomiR-NC, were introduced into cells using Lipofectamine 2000 in accordance with the manufacturer’s protocol. Luciferase activities were detected at 48 hrs post-transfection using a Dual-Luciferase® Reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blot analysis
Total protein of tissue samples or cultured cells was isolated using a Total Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Total protein concentration was examined using a BCA assay kit (Nanjing KeyGen Biotech Co., Ltd.). Equal quantities of protein were separated using 10% sodium dodecyl sulfate polyacrylamide gels and transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). Subsequently, the membranes were blocked at room temperature for 2 hrs in 5% dried skimmed milk that was dissolved in Tris-buffered saline containing 0.1% Tween-20 (TBST). After overnight incubation at 4°C with primary antibodies, the membranes were washed with TBST thrice, followed by incubation at room temperature with the goat anti-mouse (ab97023) or goat anti-rabbit (ab97051) horseradish-peroxidase-conjugated IgG secondary antibody (1:5,000 dilution; Abcam, Cambridge, UK) for 1 h. Finally, the ECL Western Blotting Analysis system (GE Healthcare, Chicago, IL, USA) was employed to visualize the protein bands. The primary antibodies used in this study included rabbit anti-human BRD4 antibody (ab128874; 1:500 dilution; Abcam), rabbit anti-human monoclonal antibody to phosphorylated phosphatidylinositol-4,5-bisphosphate 3-kinase (p-Pi3K; ab182651; 1:1,000 dilution; Abcam), mouse anti-human monoclonal PI3K antibody (ab86714; 1:1,000 dilution; Abcam), rabbit anti-human monoclonal antibody to phosphorylated protein kinase B (p-Akt; sc-81433; 1:1,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-human monoclonal Akt antibody (ab179463; 1:1,000 dilution; Abcam), and mouse anti-human GAPDH antibody (ab9484; 1:500 dilution; Abcam). GAPDH served as an internal control.
Statistical analysis
Data were presented as the means ± SD and analyzed with SPSS version 19.0 software (IBM, Armonk, New York, USA). The association between miR-612 expression and the clinicopathological factors in NSCLC patients was analyzed by the χ2 test. Spearman’s correlation analysis was used to investigate the correlation between miR-612 and BRD4 mRNA in NSCLC tissues. The difference between two groups was examined using two-tailed Student’s t-test. One-way ANOVA followed by Student–Newman–Keuls post hoc test was utilized to analyze the statistical significance between multiple groups. The prognostic value of miR-612 in patients with NSCLC was assessed with Kaplan–Meier survival analysis. The level of statistical significance was set at P<0.05.
Results
Downregulation of miR-612 is associated with poor prognosis in NSCLC
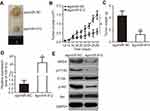
To clarify the expression profile of miR-612 in NSCLC, RT-qPCR was first performed to detect miR-612 expression in 57 pairs of NSCLC tissues and ANTs.
Results showed that miR-612 expression in NSCLC tissues was notably decreased compared with that in ANTs (Figure 1A, P<0.05). In addition, the expression differences of miR-612 in five human NSCLC cell lines (H522, H460, H1299, A549, and SK-MES-1) and non-tumorigenic bronchial epithelium BEAS-2B was investigated using RT-qPCR. Consistent with the trend observed for NSCLC tissues, the expression level of miR-612 was significantly lower in all tested NSCLC cell lines than that in BEAS-2B (Figure 1B, P<0.05).
To evaluate the clinical value of miR-612 in patients with NSCLC, all NSCLC patients were divided into either the miR-612 high expression group or low expression group based on the median value of miR-612 in NSCLC tissues. Decreased miR-612 expression was significantly correlated with tumor-node-metastasis staging system (TNM) stage (P=0.017), and lymph node metastasis (P=0.007), but not with age, sex, tumor size, or smoking (Table 1, all P>0.05). Furthermore, patients with NSCLC and lower miR-612 expression exhibited a poorer overall survival rate (Figure 1C, P=0.0064). These results suggest that downregulation of miR-612 might be closely related with the poor prognosis of patients with NSCLC.
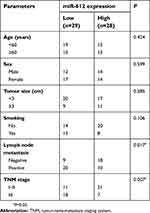 | Table 1 Associations between miR-612 expression and clinicopathological parameters in patients with NSCLC |
miR-612 inhibits the proliferation, migration, invasion and induces the apoptosis of NSCLC cells
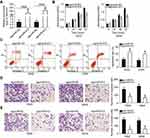
As we discovered that miR-612 is expressed at low levels in NSCLC, we inferred that miR-612 may exert tumor-suppressive roles in NSCLC cells. To this end, agomiR-612 was introduced into H522 and A549 cells to increase endogenous miR-612 expression (Figure 2A, P<0.05). Here, the influence of miR-612 overexpression on NSCLC cell proliferation was examined using the CCK-8 assay. The proliferation ability of H522 and A549 cells transfected with agomiR-612 was evidently decreased relative to that of the agomiR-NC group (Figure 2B, P<0.05). In addition, we examined the apoptosis of miR-612-overexpressing H522 and A549 cells by flow cytometry analysis. It was observed that agomiR-612 transfection significantly increased the apoptosis of H522 and A549 cells (Figure 2C, P<0.05).
Transwell migration and invasion assays were applied to explore the exact roles of miR-612 in the metastasis of NSCLC cells. The migratory (Figure 2D, P<0.05) and invasive (Figure 2E, P<0.05) capacities of H522 and A549 cells were markedly decreased following upregulation of miR-612. These results implied that miR-612 attenuated the growth and metastasis of NSCLC cells in vitro.
BRD4 is a direct target of miR-612 in NSCLC cells
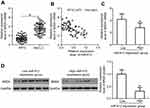
As miRNAs function by regulating the expression of their targets,9 bioinformatics analysis was utilized to predict the potential target genes of miR-612. BRD4, which contains a complementary binding site for miR-612 (Figure 3A), was selected for further identification because this gene has been reported as a regulator for the development of NSCLC.24,25 To confirm modulation of BRD4 in NSCLC cells by miR-612, RT-qPCR and western blot analysis were carried out for H522 and A549 cells transfected with agomiR-612 or agomiR-NC. The results indicated that expression levels of BRD4 mRNA (Figure 3B, P<0.05) and protein (Figure 3C, P<0.05) were downregulated in H522 and A549 cells after miR-612 upregulation. A luciferase reporter assay was further employed to explore whether the 3ʹ-UTR of BRD4 could be directly targeted by miR-612 in NSCLC cells. As shown in Figure 3D, upregulation of miR-612 in H522 and A549 cells obviously repressed the luciferase activity of the plasmid harboring the Wt miR-612 binding site (P<0.05); however, the alteration of miR-612 expression did not affect the luciferase activity of the plasmid containing the Mut binding site. In summary, these results demonstrated that BRD4 is a direct target gene of miR-612 in NSCLC cells.
miR-612 expression is inversely correlated with BRD4 expression in NSCLC tissues
We further measured BRD4 expression in NSCLC tissues and explored the expression correlation between miR-612 and BRD4. The data obtained from RT-qPCR analysis revealed that BRD4 mRNA was significantly overexpressed in NSCLC tissues compared with that in ANTs (Figure 4A, P<0.05). In addition, the mRNA level of BRD4 in NSCLC tissues exhibited a negative correlation with miR-612 expression (Figure 4B; R2=0.3470, P<0.0001). Furthermore, BRD4 mRNA (Figure 4C, P<0.05) and protein (Figure 4D, P<0.05) expression was evidently lower in the miR-612 high expression group than that in the miR-612 low expression group. These results confirmed that miR-612 expression was inversely correlated with BRD4 expression in NSCLC tissues.
Silencing BRD4 expression inhibits the growth and metastasis of NSCLC cells in vitro
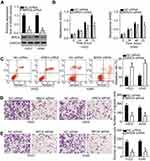
As BRD4 was identified to be a direct target of miR-612 in NSCLC cells, we next attempted to determine whether the functions of BRD4 inhibition were similar with those induced by miR-612 overexpression in NSCLC cells. BRD4 siRNA was transfected into H522 and A549 cells, and the protein level of BRD4 was efficiently knocked down in H522 and A549 cells after BRD4 siRNA transfection (Figure 5A, P<0.05). CCK-8 and flow cytometry analysis revealed that downregulation of BRD4 significantly suppressed cell proliferation (Figure 5B, P<0.05) but promoted cell apoptosis (Figure 5C, P<0.05) in H522 and A549 cells. The migration (Figure 5D, P<0.05) and invasion (Figure 5E, P<0.05) was inhibited in H522 and A549 cells upon BRD4 knockdown, as determined by Transwell migration and invasion assays. These results clearly showed that silencing BRD4 expression could imitate the tumor-suppressive roles of miR-612 upregulation in NSCLC cells, further suggesting BRD4 as a direct target gene of miR-612 in NSCLC cells.
BRD4 reverses the suppressive roles of miR-612 upregulation on the malignant phenotypes of NSCLC cells
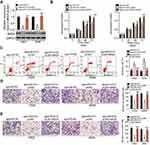
As it was revealed that miR-612 exerts a tumor-suppressive role in NSCLC progression, accompanied by identification of BRD4 as a direct target gene of miR-612 in NSCLC cells, a series of rescue experiments were applied to evaluate whether BRD4 was essential for the miR-612-mediated malignant phenotypes of NSCLC cells. BRD4 overexpression plasmid pCMV-BRD4 or empty pCMV plasmid were co-transfected with agomiR-612 into H522 and A549 cells. Western blot analysis verified that co-transfection of pCMV-BRD4 restored BRD4 protein expression in agomiR-612-transfected H522 and A549 cells (Figure 6A, P<0.05). Restoration of BRD4 expression partially abolished the influence of miR-612 overexpression on cell proliferation (Figure 6B, P<0.05), apoptosis (Figure 6C, P<0.05), migration (Figure 6D, P<0.05), and invasion (Figure 6E, P<0.05) of H522 and A549 cells. These results further demonstrated BRD4 as a direct downstream effector of miR-612 in NSCLC cells, and suppression of BRD4 is essential for the miR-612-mediated anticancer effects in NSCLC cells.
miR-612 suppresses the Pi3K/Akt signaling pathways in NSCLC cells by targeting BRD4
To further understand the exact mechanisms by which miR-612 inhibited the malignant development of NSCLC, we investigated the influence of miR-612 on important signaling pathways. As BRD4 was implicated in the regulation of the PI3K/Akt pathway,26 we explored whether this pathway could be deactivated by miR-612 overexpression in NSCLC cells. We found that transfection with agomiR-612 decreased the protein levels of p-PI3K and p-Akt in H522 and A549 cells. Notably, restored BRD4 expression could abolish the suppressive effects of miR-612 overexpression on p-PI3K and p-Akt levels in H522 and A549 cells (Figure 7). Thus, these observations suggested that miR-612 directly targeted BRD4 to inhibit the activation of PI3K/Akt pathway in NSCLC cells.
miR-612 impairs NSCLC growth in vivo through regulation of the BRD4/PI3K/Akt pathway
To further examine the tumor-suppressive action of miR-612 in NSCLC, an in vivo tumor growth assay was utilized to investigate the effect of miR-612 upregulation on tumor growth. The volume (Figure 8A and B, P<0.05) and weight (Figure 8C, P<0.05) of tumors in the agomiR-612 group were obviously lower than those in the agomiR-NC group. Next, the expression of miR-612 in the tumor xenografts was measured using RT-qPCR. The data showed that, compared with that of the agomiR-NC group, higher miR-612 expression was observed in the tumor xenografts derived from agomiR-612-transfected H522 cells (Figure 8D, P<0.05). In addition, western blot analysis indicated that the tumor xenografts obtained from the agomiR-612 group displayed decreased expression of BRD4, p-PI3K, and p-Akt in comparison with those in the agomiR-NC group (Figure 8E). These results demonstrated that upregulation of miR-612 hindered NSCLC growth in vivo. Downregulation of BRD4 and deactivation of the PI3K/Akt pathway might be responsible for the suppressive effect of miR-612 on NSCLC growth in vivo.
Discussion
Recently, the aberrant expression of miRNAs in NSCLC has been reported in various studies.27,28,29 The dysregulation of miRNAs exerts tumor-suppressing or tumor-promoting actions on the pathology biological behaviors of NSCLC.30 More importantly, miRNAs are considered to be attractive targets for anticancer therapies in patients with NSCLC.26 Thus, investigation of the expression profile and specific functions of cancer-associated miRNAs in NSCLC is critical, and it may present novel insights into the identification of promising therapeutic targets for NSCLC therapy. In the present study, for the first time, we detected the expression pattern of miR-612 in NSCLC and evaluated its clinical value in patients with NSCLC. Notably, the functional roles and regulatory mechanism of miR-612 in the aggressive behaviors of NSCLC were explored in depth. Our results revealed that miR-612 suppressed the malignant development of NSCLC in vitro and in vivo, a process that was mediated by direct targeting of BRD4 and was related to the deactivation of PI3K/Akt signaling.
miR-612 has been reported as being aberrantly expressed in multiple types of human cancer. For instance, miR-612 expression is decreased in bladder cancer, and decreased miR-612 expression is related with tumor stage, lymph node metastasis, and distant metastasis.18 miR-612 is downregulated in melanoma, and its downregulation is significantly correlated with the melanoma thickness and lymph node metastasis.19 Patients with melanoma harboring low miR-612 expression have shorter overall and disease-free survival rates.19 miR-612 is expressed at low levels in colorectal cancer and a low miR-612 expression exhibits an obvious correlation with tumor metastasis.20 miR-612 also expresses at low levels in ovarian cancer21 and endometrial carcinoma.22 However, the expression pattern and clinical significance of miR-612 in NSCLC remains poorly understood. In this study, we found that miR-612 was downregulated in NSCLC tissue specimens and a panel of cell lines. Downregulation of miR-612 was closely associated with the TNM stage and lymph node metastasis in patients with NSCLC. NSCLC patients with low miR-612 expression exhibited poorer prognosis than those patients with high miR-612 expression. These results suggest that miR-612 may serve as diagnostic and prognostic markers for patients with NSCLC.
miR-612 plays crucial roles in regulation of the malignancy. For example, exogenous miR-612 expression restricts bladder cancer cell growth, metastasis, epithelial-mesenchymal transition in vitro and tumor growth in vivo.18 Overexpression of miR-612 inhibits the proliferation, colony formation, and invasion of melanoma cells as well as impairs tumor growth in vivo.19 Resumption of miR-612 expression attenuates cell proliferation, migration, and epithelial-mesenchymal transition and decreases tumor growth in vivo in colorectal cancer.20 miR-612 also plays tumor-suppressive roles in the malignant development of NSCLC through regulating cell proliferation, colony formation, metastasis, epithelial-mesenchymal transition, stem cell-like property, and chemoresistance.31,32,33 However, the biological functions of miR-612 in NSCLC progression remain largely unclear. Herein, a series of functional experiments revealed that miR-612 overexpression inhibited NSCLC cell proliferation, migration, and invasion in vitro, promoted cell apoptosis in vitro, and reduced the tumor growth in vivo. These results demonstrated that miR-612 functions as a tumor-suppressive miRNA that exerts important regulatory roles in the occurrence and development of NSCLC, suggesting miR-612 as a novel therapeutic target for NSCLC.
Regarding the underlying mechanisms, multiple genes, including malic enzyme 1,18 espin,19 AKT2,20 specificity protein,133 and HOXA13,21 have been confirmed as direct targets of miR-612. In the present study, we explored the exact molecular mechanisms by which miR-612 suppressed tumor processes in NSCLC. BRD4, a member of the bromodomain and extra-terminal domain (BET) family,34 was demonstrated to be a novel direct target of miR-612 in NSCLC cells. BRD4 is expressed at high levels in NSCLC, and high BRD4 expression exhibits a significant correlation with histological type, lymph node metastasis, tumor stage, and differentiation.24 Patients with NSCLC with high BRD4 expression have a poorer prognosis than patients with low BRD4 expression.24 BRD4 exerts a critical role in negatively regulating NSCLC cell growth, apoptosis, migration, and invasion.24,25 Here, we demonstrated that miR-612 inhibited BRD4 expression by directly targeting its 3ʹ-UTR and subsequently repressed the activation of the PI3K/Akt pathway, resulting in the suppression of malignant phenotypes of NSCLC cells in vitro and in vivo. Accordingly, silencing BRD4 and deactivating the PI3K/Akt pathway using miR-612-mediated target therapy might be a potential therapeutic approach for managing patients with NSCLC.
Conclusion
In conclusion, this study revealed that miR-612 expression was decreased in NSCLC and low expression predicted a poor survival of patients with NSCLC. Exogenous miR-612 expression suppressed NSCLC cell proliferation, migration, and invasion, and promoted cell apoptosis through direct targeting of BRD4 and deactivation of the PI3K/Akt pathway. Thus, these observations suggest that miR-612 acts as a tumor-suppressive miRNA in NSCLC and might be a promising target for NSCLC therapy. However, the sample size of this study was small. In addition, we did not use antagomiR-612 to silence endogenous miR-612 expression and explored the influence of miR-612 knockdown on malignancy of NSCLC cells. We will resolve the two limitations in the near future.
Acknowledgments
This study was supported by the Heilongjiang Postdoctoral Science Foundation (LBH-Z16110), Wu Jieping Medical Foundation Clinical Research Fund (320.6750.17138), and China Postdoctoral Science Foundation (2017M610213).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi:10.3322/caac.21262
2. Li X, Zhang Q, Fan K, et al. Overexpression of TRPV3 correlates with tumor progression in non-small cell lung cancer. Int J Mol Sci. 2016;17:437.
3. Harada H, Murayama S. Proton beam therapy in non-small cell lung cancer: state of the art. Lung Cancer. 2017;8:141–145. doi:10.2147/LCTT.S117647
4. Ettinger DS, Akerley W, Borghaei H, et al. Nccn Non-small cell lung cancer. J Natl Compr Cancer Network. 2012;10:1236–1271.
5. Boffetta P, Nyberg F. Contribution of environmental factors to cancer risk. Br Med Bull. 2003;68:71–94.
6. Ridge CA, McErlean AM, Ginsberg MS. Epidemiology of lung cancer. Semin Intervent Radiol. 2013;30:93–98. doi:10.1055/s-0033-1342949
7. de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018;7:220–233. doi:10.21037/tlcr.2018.05.06
8. Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. doi:10.12688/f1000research
9. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
10. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297.
11. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi:10.1016/j.cell.2009.01.002
12. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96 Suppl:R40–44.
13. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi:10.1038/nrc2867
14. Wang Y, Yu L, Wang T. MicroRNA-374b inhibits the tumor growth and promotes apoptosis in non-small cell lung cancer tissue through the p38/ERK signaling pathway by targeting JAM-2. J Thorac Dis. 2018;10:5489–5498. doi:10.21037/jtd.2018.09.93
15. Jiang M, Zhou LY, Xu N, An Q. Down-regulation of miR-500 and miR-628 suppress non-small cell lung cancer proliferation, migration and invasion by targeting ING1. Biomed Pharmacother. 2018;108:1628–1639. doi:10.1016/j.biopha.2018.09.145
16. Zhang YS, Chen T, Cai YJ, et al. MicroRNA-647 promotes the therapeutic effectiveness of argon-helium cryoablation and inhibits cell proliferation through targeting TRAF2 via the NF-kappaB signaling pathway in non-small cell lung cancer. Onco Targets Ther. 2018;11:6777–6784. doi:10.2147/OTT.S159337
17. Lamichhane SR, Thachil T, De Ieso P, Gee H, Moss SA, Milic N. Prognostic role of microRNAs in human non-small-cell lung cancer: a systematic review and meta-analysis. Dis Markers. 2018;2018:8309015. doi:10.1155/2018/8309015
18. Liu M, Chen Y, Huang B, et al. Tumor-suppressing effects of microRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. Int J Oncol. 2018;52:1923–1933. doi:10.3892/ijo.2018.4342
19. Zhu Y, Zhang HL, Wang QY, Chen MJ, Liu LB. Overexpression of microRNA-612 restrains the growth, invasion, and tumorigenesis of melanoma cells by targeting Espin. Mol Cell. 2018;41:119–126. doi:10.14348/molcells.2018.2235
20. Sheng L, He P, Yang X, Zhou M, Feng Q. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis. 2015;6:e1808. doi:10.1038/cddis.2015.184
21. Yu H, Xu Y, Zhang D, Liu G. Long noncoding RNA LUCAT1 promotes malignancy of ovarian cancer through regulation of miR-612/HOXA13 pathway. Biochem Biophys Res Commun. 2018;503:2095–2100. doi:10.1016/j.bbrc.2018.07.165
22. Zhang L, Wang DL, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22:4820–4827. doi:10.26355/eurrev_201808_15617
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
24. Liao YF, Wu YB, Long X, et al. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491–9500. doi:10.18632/oncotarget.7068
25. Gao Z, Yuan T, Zhou X, et al. Targeting BRD4 proteins suppresses the growth of NSCLC through downregulation of eIF4E expression. Cancer Biol Ther. 2018;19:407–415. doi:10.1080/15384047.2018.1423923
26. Boeri M, Pastorino U, Sozzi G. Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J. 2012;18:268–274. doi:10.1097/PPO.0b013e318258b743
27. Jiang W, He Y, Shi Y, et al. MicroRNA-1204 promotes cell proliferation by regulating PITX1 in non-small cell lung cancer. Cell Biol Int. 2018. doi:10.1002/cbin.11083
28. Xie Q, Yu Z, Lu Y, Fan J, Ni Y, Ma L. microRNA-148a-3p inhibited the proliferation and epithelial-mesenchymal transition progression of non-small-cell lung cancer via modulating Ras/MAPK/Erk signaling. J Cell Physiol. 2018. doi:10.1002/jcp.27899
29. Bagheri A, Khorshid HRK, Tavallaie M, et al. A panel of noncoding RNAs in non-small-cell lung cancer. J Cell Biochem. Epub 2018 Nov 28.
30. Gao W, Xu J, Shu YQ. miRNA expression and its clinical implications for the prevention and diagnosis of non-small-cell lung cancer. Expert Rev Respir Med. 2011;5:699–709. doi:10.1586/ers.11.55
31. Tao ZH, Wan JL, Zeng LY, et al. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi:10.1084/jem.20120153
32. Tang J, Tao ZH, Wen D, et al. MiR-612 suppresses the stemness of liver cancer via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;447:210–215. doi:10.1016/j.bbrc.2014.03.135
33. Liu Y, Liu DL, Dong LL, et al. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis. 2016;7:e2377. doi:10.1038/cddis.2016.282
34. Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi:10.1038/nrc3256
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.