Back to Journals » Neuropsychiatric Disease and Treatment » Volume 16
MicroRNA-382-5p Targets Nuclear Receptor Subfamily 3 Group C Member 1 to Regulate Depressive-Like Behaviors Induced by Chronic Unpredictable Mild Stress in Rats
Authors Li S, Ma H, Yuan X, Zhou X, Wan Y, Chen S
Received 27 December 2019
Accepted for publication 17 August 2020
Published 9 September 2020 Volume 2020:16 Pages 2053—2061
DOI https://doi.org/10.2147/NDT.S243920
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Shuqian Li,1 Hong Ma,2 Xu Yuan,1 Xiaomei Zhou,1 Yiwen Wan,1 Shangjie Chen1
1Department of Rehabilitation, People’s Hospital of Shenzhen Baoan District, Shenzhen 518100, People’s Republic of China; 2Department of Rehabilitation, Binzhou Medical University, Yantai, Shandong Province 264003, People’s Republic of China
Correspondence: Shangjie Chen
Department of Rehabilitation, People’s Hospital of Shenzhen Baoan District, 118 Longjing Second Road, Xin’an Street, Shenzhen, Baoan District 518100, People’s Republic of China
Email [email protected]
Background: Depression is an emotional disorder characterized by depression, lack of pleasure, and cognitive and sleep disorders. It is a systemic disease with a complex pathogenesis. In this study, we will be focused to investigate their associations and the exact functional mechanisms of miR-382-5p and NR3C1 in depression.
Materials and Methods: We measured the expressions of microRNA-382-5p (miR-382-5p) and NR3C1 in the hippocampus by chronic unpredictable mild stress (CUMS). Depression behavior test including novelty-suppressed feeding test (NSFT), sucrose preference test (SPT), and forced swim test (FST) on rats have been conducted to examine the roles and functions of miR-382-5p and NR3C1 on depression-like behaviors by lentivirus vectors.
Results: Up-regulation of miR-382-5p and down-regulation of NR3C1 were observed in rats’ hippocampus induced by CUMS. miR-382-5p targeted NR3C1 and inhibited the expressions of NR3C1 in rats’ hippocampus. miR-382-5p could significantly change the depression behaviors induced by CUMS. NR3C1 downstream BDNF and p-TrkB were also oppositely associated with miR-382-5p in rats’ hippocampus.
Conclusion: Through our experiments and analysis, we found that the associations between miR-382-5p and NR3C1 could affect the depression-like behaviors.
Keywords: miRNA-382-5p, depression, hippocampus, NR3C1
Background
Depression is an emotional disorder characterized by a lack of pleasure, cognitive, and sleep disorders.1,2 It is a systemic disease with a complex pathogenesis.3,4 Compared with the current first-line treatments targeting monoamine transmitters, the development of drugs that has a therapeutic impact on multiple pathogenesis is a new topic.5 In recent years, studies have shown that endogenous non-coding small RNAs with regulatory functions are abnormally expressed in tissues and body fluids of animal models and depressed patients, and in drug therapy response, synaptic plasticity, neuron production, cognition, learning and memory functions.6,7 Among them, microRNAs-based transcriptomics studies have made progress in the discovery of clinical depression biomarkers and potential drug targets.8
MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs found in eukaryotes that are about 22 nt in length and have regulatory functions,9 which are able to complement the 3ʹ non-coding region of target genes by sequence complementation.10,11 Accumulative evidence suggest that miRNA is involved in multiple classical pathogenesis of depression, by targeting multiple nodal genes, thereby regulating the expressions of key proteins, which means that miRNA itself has great potential as a therapeutic target for depression and a prospective biomarker.12,13 In a recent report, Lopozzp et al discussed that miRNAs in depression-like behaviors,14 which shed light to the potential of utilizing many miRNAs in the development or treatment of depression. It has been reported that several miRNAs including miR-9-5p, miR-128-1-5p, and miR-382-5p that are significantly increased in chronic mild stress.15 As an important member of miRNAs, miR-382-5p has been demonstrated to play important roles in liver cancer metastasis,16 ischemic stroke,17 and stem cell differentiation.18 In 2015, RJ. Chen reported that miR-382-5p could act as a biomarker of resilience or vulnerability to stress,19 which gives us a clue in the depression. Herein, we are interested in the exact role of miR-382-5p in depression behaviors.
Hippocampus has been considered as a core factor in the pathophysiology of depression, and previous studies have revealed that hippocampal glucocorticoid receptor gene nuclear receptor subfamily 3 group C member 1 (NR3C1) DNA methylation can mediate part of preconception paternal stress effects in rat offspring.20 It was also revealed that there exist genetic and epigenetic associations of MAOA and NR3C1 with depression.21 Besides, NR3C1 promoter was proved to be hypermethylated in Thai females with major depressive disorder.22 The preliminary bioinformatics analysis revealed that miR-382-5p could target NR3C1. We hypothesis that miR-382-5p may exert a modulation role by interacting with NR3C1 in depression. Therefore, we will be focused to investigate their associations and the exact functional mechanisms.
Materials and Methods
Experimental Animals
Male Wistar rats with weights at around 200 grams were provided by Shanghai Animal Center, China. They were maintained in half day light and half day night at ~22Celsius and 55% humidity, with freedom to eat and drink. The experiments had been approved by the Institute for Experimental Animals of People’s Hospital of Shenzhen Baoan District, and our study followed the guidelines of the National Institutes of Health.
Chronic Unpredictable Mild Stress (CUMS) Rats
Rats in CUMS groups had various stress factors, such as 1 day’s starvation, 1 day’s water deprivation, 5 minutes’ cold swimming, and 1 minute’s tail pinch randomly for 28 days. Control group was kept in a separate place. The rats were given behavioral tests before being sacrificed. For euthanasia, rats were anesthetized by intraperitoneal injection with 3% sodium pentobarbital (30 mg/kg body weight, Sigma Chemical Co., St. Louis, MO, USA) and sacrificed by cervical dislocation.
Lentivirus Vectors and Stereotaxic Injection
Construction of NR3C1 lentiviral vector (LV-NR3C1) and subsequent production of lentivirus were completed by Gene Pharma Company (Shanghai, China). The empty vector (LV-Mock) served as the negative control. LV-miR-382-5p/LV-si-miR-382-5p (108 TU/mL) also purchased from the Gene Pharma Company (Shanghai, China). All viral injection procedures followed the previous study.23 For stereotaxic surgery test, rats were anesthetized with a 100 mg/kg ketamine and 10 mg/kg xylazine mixture and installed in a stereotaxic framework. One milliliter of the virus solution was bilaterally infused into rats using a precision Hamilton micro-syringe with a needle of 26 G, according to the following coordinates: hippocampus [The first injection: 4.8 mm posterior from Bregma, 2.5 mm lateral from the medial suture, 3.5 mm ventral to the surface of the skull. The second injection: 4.8 mm posterior from Bregma, 5 mm lateral from the medial suture, 6 mm ventral to surface of the skull]. Rats recovered 1 week before exposure to CUMS.
Depression Behavioral Test
Novelty-suppressed feeding test (NSFT): the NSFT was performed 4 weeks after CUMS exposure. The test instrument consisted of a plastic box (35 cm x 35 cm). No food was given to the rats 24 h before the test. At the start of the test, a single pellet of food was put on a white paper platform in the middle of the box. We put a rat in a corner of the maze and immediately activated a stopwatch. Interest test was started when the rats grabbed the food with their forepaws and began to eat. As a control value, we measured the food consumption in home cages within 90 min after the completion of the test; Sucrose preference test (SPT): Rats were trained to adapt to a sucrose solution (1%, w/v) before this test. Two bottles of sucrose solution were placed for 24 h in an individual cage, and then one bottle of sucrose solution was changed into water for 24 h. After adaptation, rats were fasted on water and food for 24 h. During the test, rats were kept in separate cages and were free to take two bottles of sucrose solution and water. The sucrose solution and water consumption were measured after 24 h; Forced swim test (FST): the FST was performed 24 h after SPT. Firstly, pour tap water (22–26°C) into glass beakers. Rats were then put into that glass beakers, where they could not escape or rest though touching the bottom of the beaker. The test lasted 6 min (360 s) and recorded the duration of immobility. Mobility was defined as any action beyond what was necessary to keep the head above water. Data were expressed in the term of the average time of immobility, swimming and climbing over the observation period (360 s).
Primary Hippocampal Neuronal Cultures
To start, we took the hippocampi rat’s brain. The neuronal cells were digested, re-suspended, and diluted to 100 cells/mL. Three-milliliter cell suspensions were cultured for 2 h in 5% CO2 at 37 Celsius for 14 days, with medium refreshed with 3 mL of neurobasal/B27 medium. After cultured neurons were treated with LV-miR-382-5p or LV-si-miR-382-5p for 48 h, protein expression levels of NR3C1, BDNF and TrkB were detected by Western blot.
qRT-PCR
Hippocampus was dissected and stored at −80 Celsius. RNA was extracted by TRIzol (Invitrogen, USA), and cDNA was generated by PrimeScript® (Takara, China). qRT-PCR was carried out to measure miRNAs and mRNA expressions by SYBR II (Takara, China). The relative expressions were calculated by 2−ΔΔCT, using U6 snRNA and GAPDH as references. The primers are displayed in Table 1.
 |
Table 1 Sequences of Primers Used in qRT-PCR |
Western Blotting
Proteins were separated by SDS-PAGE and transferred to PVDF membrane (Bio-Rad, USA). We blocked the membranes and incubated them with anti-NR3C1 (1:500, sc-12763), anti-BDNF (1:500, sc-546), anti-TrkB (1:500, sc-7268), p-TrkB (1:500, sc-8058), which were purchased from Santa Cruz Biotechnology, USA. GAPDH (1:1000, A9044) from Sigma, USA was used as control. After washing, we incubated the blots with HRP-conjugated secondary antibodies. Chemiluminescent was used to quantify the signals.
Luciferase Assay
The possible target locations of the NR3C1 3′-untranslated region (3′-UTR) (WT-NR3C1) or mutated sequences (MT-NR3C1) were cloned into the pmirGLO Dual-Luciferase miRNA target expression vector (Promega, Madison, WI). The total 3′-UTR NR3C1 sequences (SwitchDB, Menlo Park, CA) were constructed using the transfection-ready luciferase reporter. Luciferase assays were operated in 293 T cells by the Dual-Luciferase Reporter 1000 Assay System (Promega). Using Normalized values (firefly activity/Renilla activity) for analysis.24
Statistical Analysis
SPSS 19.0 was used for data analysis. Data were shown as means ± SD. The difference significance was determined by unpaired t-test (2 groups) or one-way ANOVA (>2 groups). Two-sided P values less than 0.05 were considered as statistically significant.
Results
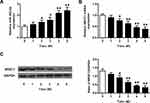
Up-Regulation of miR-382-5p and Down-Regulation of NR3C1 in Rats’ Hippocampus Induced Through CUMS
We assessed the transcriptional and translational level of miR-382-5p and NR3C1 in rats’ hippocampus to investigate the expression level of miR-382-5p in CUMS model. As shown in Figure 1A, CUMS induced miR-382-5p expressions, which increased further with time passing by (n=6 per group. P<0.05, P<0.01). Differently, Figure 1B and C showed that hippocampus NR3C1 was down-regulated in CUMS rats as time passing by (P<0.05, P<0.01).
Impact of miR-382-5p Over-Expressions on the Behaviors Induced by CUMS
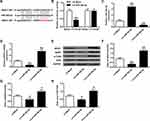
In contrast with rats transfected with LV-Mock, rats with miR-382-5p over-expressions had elevated latency to feed (P<0.01) (Figure 2A) and immobility time (P<0.01) (Figure 2E), and lower sucrose preference (P<0.01) (Figure 2C) and swimming time (P<0.01) (Figure 2F), which suggested more depression. Inhibition of miR-382-5p had the opposite effects. Food intake (Figure 2B) and total water intake (Figure 2D) did not show any significant change. Our results indicated that miR-382-5p can significantly enhance the CUMS induced depression behaviors.
miR-382-5p Down-Regulates the Expressions of NR3C1 in Rats’ Hippocampus
To evaluate the influence of miR-382-5p on the expression of NR3C1, we conducted gain and loss function experiments. Figure 3A finds decreased or enhanced expressions of miR-382-5p in rats’ hippocampus by LV-si-miR-382-5p and LV-miR-382-5p (P<0.01). Figure 3B–D revealed that NR3C1 expressions were significantly lowered in miR-382-5p and elevated in si-miR-382-5p (P<0.01). Figure 3E and F indicated that the NR3C1 downstream BDNF and p-TrkB were also oppositely associated with miR-382-5p in rats’ hippocampus (P<0.05).
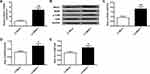
NR3C1 is Targeted by miR-382-5p
Further investigation of miR-382-5p and NR3C1 were conducted. As shown in Figure 4A, bioinformatics used to predict that NR3C1 was a target of miR-382-5p. Figure 4B uses luciferase reporter assays and found that NR3C1-mut abolished its abilities to bind with the miR-382-5p 3′-UTR (P>0.05). Figure 4C shows cultured neuronal cells were treated with LV-miR-382-5p or LV-si-miR-382-5p (P<0.01). Figure 4D–H demonstrated that NR3C1, BDNF and p-TrkB were elevated with LV-si-miR-382-5p treatments and lowered with LV-miR-382-5p (P<0.05, P<0.01). From these results, we could conclude that NR3C1 is targeted by miR-382-5p.
NR3C1 Over-Expressions on CUMS-Related Depression Behaviors in Rats
To further illustrate the function of NR3C1 over-expression on CUMS, we analyzed the level of NR3C1 in rats. According to the qPCR and Western blotting results, we found that NR3C1 expressions were significantly elevated under LV-NR3C1 (P<0.01) (Figure 5A–C). Figure 5B, D, and E demonstrates that NR3C1 downstream targets BDNF and p-TrkB were also elevated in the LV-NR3C1 group (P<0.05).
Then, the depression behaviors of Rats described in previous studies25,26 were also assessed in the delay in the latency to feed (Figure 6A), food intake (Figure 6B), sucrose preference (Figure 6C), total water intake (Figure 6D), immobility time (Figure 6E), and swimming time (Figure 6F) under treatments of Control, CUMS, CUMS+LV-Mock, CUMS+LV-NR3C1. Treatments with LV-NR3C1 lowered the latency to feed (P<0.05), elevated sucrose preference (P<0.01), decreased immobility time (P<0.05), and elevated swimming time (P<0.01). No obvious changes were observed in food intake and water intake by NR3C1 over-expressions.
Discussions
MiRNAs have specific expressions in neural progenitor cells, neurons and glial cells during neurodevelopment. For instance, researchers have revealed that miR-1202,27 miR-137,28 and miR-144-5p29 may be involved in physiological processes such as axon guidance, synaptic plasticity regulation, and neurodegenerative diseases and depression.30 From our study, we found that CUMS induced the promotion of miR-382-5p expressions in a dose-dependent manner. Differently, hippocampus NR3C1 was lowered by CUMS with time passing by. It was obvious that miR-382-5p was up-regulated and NR3C1 was down-regulated in the hippocampus of rats induced by CUMS. From these data, we believe that alteration of miR-382-5p and NR3C1 in the hippocampus may be related to stress responses in rats.
Previous studies have demonstrated that miRNAs played an important role in neuronal development and differentiation, synaptic plasticity, cognition and learning and memory functions.31,32 Some miRNAs are associated with the development of various central nervous system diseases.7,33 In contrast with rats transfected with LV-Mock, rats with miR-382-5p over-expressions had elevated latency to feed and immobility time, and lower sucrose preference and swimming time, which suggested more depression. Inhibition of miR-382-5p had the opposite effects. No significant change was found in food intake and total water intake.
It has been reported that depression factors such as childhood life unfortunate could affect the methylation level of the NR3C1 promoter region.34,35 The methylation level of the NR3C1 promoter region in patients with depression was positively correlated with the hippocampal subdivision volume.36 In our experiments, we found decreased or enhanced expressions of miR-382-5p in rats’ hippocampus by LV-si-miR-382-5p and LV-miR-382-5p. NR3C1 expressions were significantly lower in miR-382-5p and elevated in si-miR-382-5p. NR3C1 downstream BDNF and p-TrkB were also oppositely associated with miR-382-5p in rats’ hippocampus. miR-382-5p inhibited the expressions of NR3C1 in rats’ hippocampus, which proved the roles of miR-382-5p in depression.
Bioinformatics analysis predicted that NR3C1 was a target of miR-382-5p. Luciferase reporter assays found that NR3C1-mut abolished its abilities to bind with the miR-382-5p 3′-UTR. NR3C1, BDNF and p-TrkB were elevated with LV-si-miR-382-5p treatments and lowered with LV-miR-382-5p. According to established results, miRNAs can also directly affect the expressions and regulation of several other key components of depression-related signaling pathways, such as brain-derived neurotrophic factor (BDNF), which is reduced in depression patients and stress animal models.1,35 Our data supported that NR3C1 was targeted by miR-382-5p, which could help modulate the levels of depression-related factors.
NR3C1 has been reported essential in the depressive disorders for a long time.37 According to the qPCR and Western blotting results, NR3C1 downstream targets BDNF and p-TrkB were also elevated in the LV-NR3C1 group. Treatments with LV-NR3C1 lowered the latency to feed, elevated sucrose preference, decreased immobility time and elevated swimming time. No obvious changes were observed in food intake and water intake by NR3C1 over-expressions. In consistence with previous findings, we also found that NR3C1 could impact depression behaviors in rats.
Conclusion
Through our experiments and analysis, we found that the associations between miR-382-5p on NR3C1 could affect the depression-like behaviors.
Abbreviations
CUMS, chronic unpredictable mild stress; NR3C1, nuclear receptor subfamily 3 group C member 1.
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The experiments had approval by the Institute for Experimental Animals of People’s Hospital of Shenzhen Baoan District.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Murphy GM, Sarginson JE, Ryan HS, O’Hara R, Schatzberg AF, Lazzeroni LC. BDNF and CREB1 genetic variants interact to affect antidepressant treatment outcomes in geriatric depression. Pharmacogenet Genomics. 2013;23(6):301–313. doi:10.1097/FPC.0b013e328360b175
2. Blazer I, German D. Depression in Late Life. CV Mosby Co; 1993.
3. Bibring E. The mechanism of depression. 1953.
4. Faulstich M. Depression–Pediatric. Psychiatry. 1986;143:1024–1027.
5. Katon W, Sullivan MD. Depression and chronic medical illness. J Clin Psychiatry. 1990;51(Suppl 6):3–11.
6. Dalton VS, Kolshus E, McLoughlin DM. Epigenetics and depression: return of the repressed. J Affect Disord. 2014;155:1–12. doi:10.1016/j.jad.2013.10.028
7. Ma K, Guo L, Xu A, Cui S, Wang J-H. Molecular mechanism for stress-induced depression assessed by sequencing miRNA and mRNA in medial prefrontal cortex. PLoS One. 2016;11(7):e0159093. doi:10.1371/journal.pone.0159093
8. Li Y-J, Xu M, Gao Z-H, et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One. 2013;8(5):e63648. doi:10.1371/journal.pone.0063648
9. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci. 2014;16(1):43.
10. Gururajan A, Naughton M, Scott KA, et al. MicroRNAs as biomarkers for major depression: a role for let-7b and let-7c. Transl Psychiatry. 2016;6(8):e862–e. doi:10.1038/tp.2016.131
11. Biggar KK, Storey KB. The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol. 2010;3(3):167–175.
12. Maffioletti E, Cattaneo A, Rosso G, et al. Peripheral whole blood microRNA alterations in major depression and bipolar disorder. J Affect Disord. 2016;200:250–258. doi:10.1016/j.jad.2016.04.021
13. Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One. 2014;9(1):e86469. doi:10.1371/journal.pone.0086469
14. Lopizzo N, Zonca V, Cattane N, Pariante CM, Cattaneo A. miRNAs in depression vulnerability and resilience: novel targets for preventive strategies. J Neural Transm. 2019;126(9):1241–1258.
15. Buran I, Etem EO, Tektemur A, Elyas H. Treatment with TREK1 and TRPC3/6 ion channel inhibitors upregulates microRNA expression in a mouse model of chronic mild stress. Neurosci Lett. 2017;656:51–57. doi:10.1016/j.neulet.2017.07.017
16. Du J, Bai F, Zhao P, et al. Hepatitis B core protein promotes liver cancer metastasis through miR-382-5p/DLC-1 axis. Biochimica Et Biophysica Acta Mol Cell Res. 2018;1865(1):1–11. doi:10.1016/j.bbamcr.2017.09.020
17. Wang Y, Ma Z, Kan P, Zhang B. The diagnostic value of serum miRNA-221-3p, miRNA-382-5p, and miRNA-4271 in ischemic stroke. J Stroke Cerebrovasc Dis. 2017;26(5):1055–1060. doi:10.1016/j.jstrokecerebrovasdis.2016.12.019
18. Zini R, Rossi C, Norfo R, et al. miR-382-5p controls hematopoietic stem cell differentiation through the downregulation of MXD1. Stem Cells Dev. 2016;25(19):1433–1443. doi:10.1089/scd.2016.0150
19. Chen R, Kelly G, Sengupta A, et al. MicroRNAs as biomarkers of resilience or vulnerability to stress. Neuroscience. 2015;305:36–48. doi:10.1016/j.neuroscience.2015.07.045
20. Niknazar S, Nahavandi A, Peyvandi AA, Peyvandi H, Roozbahany NA, Abbaszadeh H-A. Hippocampal NR3C1 DNA methylation can mediate part of preconception paternal stress effects in rat offspring. Behav Brain Res. 2017;324:71–76. doi:10.1016/j.bbr.2017.02.014
21. Melas PA, Wei Y, Wong CC, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int J Neuropsychopharm. 2013;16(7):1513–1528. doi:10.1017/S1461145713000102
22. Nantharat M, Wanitchanon T, Amesbutr M, Tammachote R, Praphanphoj V. Glucocorticoid receptor gene (NR3C1) promoter is hypermethylated in Thai females with major depressive disorder. Gene Mol Res. 2015;14(4):19071–19079. doi:10.4238/2015.December.29.15
23. Bahi A, Chandrasekar V, Dreyer J-L. Selective lentiviral-mediated suppression of microRNA124a in the hippocampus evokes antidepressants-like effects in rats. Psychoneuroendocrinology. 2014;46:78–87. doi:10.1016/j.psyneuen.2014.04.009
24. Lee ST, Chu K, Jung KH, et al. miR-206 regulates brain‐derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–277. doi:10.1002/ana.23588
25. Sasibhushana RB, Shankaranarayana Rao BS, Srikumar BN. Repeated finasteride administration induces depression-like behavior in adult male rats. Behav Brain Res. 2019;365:185–189. doi:10.1016/j.bbr.2019.03.006
26. Tang MM, Lin WJ, Pan YQ, Guan XT, Li YC. Hippocampal neurogenesis dysfunction linked to depressive-like behaviors in a neuroinflammation induced model of depression. Physiol Behav. 2016;161:166–173. doi:10.1016/j.physbeh.2016.04.034
27. Lopez JP, Lim R, Cruceanu C, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med. 2014;20(7):764. doi:10.1038/nm.3582
28. Zhao L, Li H, Guo R, et al. miR-137, a new target for post-stroke depression? Neural Regeneration Res. 2013;8(26):2441.
29. Wang X, Sundquist K, Hedelius A, Palmér K, Memon AA, Sundquist J. Circulating microRNA-144-5p is associated with depressive disorders. Clin Epigenetics. 2015;7(1):69. doi:10.1186/s13148-015-0099-8
30. Ha T-Y. MicroRNAs in human diseases: from autoimmune diseases to skin, psychiatric and neurodegenerative diseases. Immune Netw. 2011;11(5):227–244. doi:10.4110/in.2011.11.5.227
31. Mouillet-Richard S, Baudry A, Launay J-M KO, Kellermann O. MicroRNAs and depression. Neurobiol Dis. 2012;46(2):272–278. doi:10.1016/j.nbd.2011.12.035
32. Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharm. 2013;23(7):602–611. doi:10.1016/j.euroneuro.2012.06.013
33. Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One. 2012;7(3):e33201. doi:10.1371/journal.pone.0033201
34. Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi:10.4161/epi.3.2.6034
35. Braithwaite E, Kundakovic M, Ramchandani P, Murphy S, Champagne F. Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics. 2015;10(5):408–417. doi:10.1080/15592294.2015.1039221
36. Na K-S, Chang HS, Won E, et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One. 2014;9(1):
37. van West D, Van Den Eede F, Del-Favero J, et al. Glucocorticoid receptor gene-based snp analysis in patients with recurrent major depression. Neuropsychopharmacol. 2006;31(3):620–627. doi:10.1038/sj.npp.1300898
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.