Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-3651 promotes the growth and invasion of hepatocellular carcinoma cells by targeting PTEN
Authors Zhao X, Song Q, Miao G, Zhu X
Received 27 April 2019
Accepted for publication 15 August 2019
Published 29 August 2019 Volume 2019:12 Pages 7045—7054
DOI https://doi.org/10.2147/OTT.S213705
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Xinyang Zhao,1,* Qilong Song,2,* Ge Miao,3,* Xinfeng Zhu1
1Department of Hepatobiliary Surgery, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, People’s Republic of China; 2Department of Gastroenterology, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, People’s Republic of China; 3Department of Outpatient Guidance, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, People’s Republic of China
Correspondence: Xinfeng Zhu
Department of Hepatobiliary Surgery, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology, No. 26 Shengli Street, Jiang’an District, Wuhan, Hubei 430030, People’s Republic of China
Email [email protected]
*These authors contributed equally to this work
Background: Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in human worldwide. Evidence indicated that upregulation of microRNA-3651 (miR-3651) was observed in human HCC tissues. In this study, we explored the mechanisms by which miR-3651 regulated the proliferation, apoptosis and invasion of HCC.
Methods: The levels of miR-3651 in human HCC tissues were detected using qRT-PCR assay. In addition, transwell invasion and Western blot assay were conducted to detect cell invasion and apoptosis, respectively. Meanwhile, the dual-luciferase reporter assay was used to explore the interaction of miR-3651 and phosphate and tension homology deleted on chromsome ten (PTEN) in HCC.
Results: The levels of miR-3651 were upregulated in HCC tissues in comparison with the matched normal tissues. Overexpression of miR-3651 significantly promoted the proliferation and invasion of Huh-7 cells. In contrast, inhibition of miR-3651 markedly inhibited the proliferation and invasion of Huh-7 cells via promoting apoptosis. Moreover, downregulation of miR-3651 markedly inhibited tumor growth in vivo. Furthermore, bioinformatics analysis and luciferase reporter assay identified that PTEN was the directly binding target of miR-3651 in Huh-7 cells. Meanwhile, overexpression of miR-3651 obviously decreased the level of PTEN, and increased the expressions of p-p85 and p-Akt in Huh-7 cells.
Conclusion: These results indicated that miR-3651 might act as a potential oncogene in HCC by targeting PTEN. Therefore, miR-3651 might be a novel therapeutic target for the treatment of HCC.
Keywords: hepatocellular carcinoma, microRNA-3651, PTEN, apoptosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common primary malignant human tumors worldwide.1,2 Hepatitis B virus and hepatitis C virus are the vital etiological factors, which could lead to cirrhosis, hepatic failure and HCC.3,4 HCC is characterized by rapid development of metastasis and fast progression.5 Surgical resection, regional ablation and liver transplantation are available for HCC treatment, whereas these therapeutic methods generally lead to poor prognosis of patients with HCC.6,7 In addition, a large rate of patients have a high recurrence rate and a poor 5-year survival rate after surgical resection.8 Although several therapies options have been applied in the clinical practice, the prognosis of HCC has obviously not improved.9 Therefore, it is important to search novel therapeutic target for HCC.
MicroRNAs (miRNAs), a small non-coding RNAs, which could bind to the 3′ untranslated regions (3’UTRs) of target mRNAs.10 It has been shown that miRNAs were involved in multiple biological processes, such as cell growth, apoptosis, metabolism and differentiation.11 Currently, miRNAs have been identified to be associated with the development and progression of HCC.12,13 Recent reports indicated that miR-3651 played vital roles in tumorigenesis and cancer progression.14,15 miRNA microarray analysis revealed that the level of miR-3651 was significantly upregulated in HCC.16 Therefore, it was interesting that whether miR-3651 could be the key factor in HCC tumorigenesis. Nevertheless, the biological effect of miR-3651 in HCC remains unclear. Therefore, this study aimed to investigate the function of miR-3651 in HCC tumorigenesis.
Materials and methods
Cell culture
Human HCC cell line Huh-7 was purchased from American Type Culture Collection (Rockville, MD, USA). Human HCC cell line Bel-7402 was obtained from Shanghai Institutes for Biological Sciences, CAS (Shanghai, People’s Republic of China). Huh-7 and Bel-7402 cells were cultured in RPMI 1640 medium (Gibco, People’s Republic of China) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin and 100 U/mL streptomycin in an incubator with 5% CO2 at 37°C.
Clinical specimens
A total of 30 paired HCC tissues and matched adjacent noncancerous tissues were obtained from 30 patients diagnosed with HCC. The patients included 20 males and 10 females, the median age of 51, range 45–64 years. None of the HCC patients received preoperative interventional therapy. This study was approved by the Institutional Ethics Committee of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. Written informed consent was obtained from all the patients. This study was conducted in accordance with the Declaration of Helsinki.
Lentiviral construction and cell transfection
The PTEN-shRNA plasmid was obtained from Thermo Fisher Scientific. 293T cells were infected with PTEN-shRNA lentivirus. After 72 hrs of infection, the supernatant containing the retroviral particles was collected. Huh7 cells were infected with lenti-vector (NC), or PTEN-shRNA supernatant for 24 hrs, and then cells were treated with puromycin (2.5 μg/mL, Thermo Fisher Scientific) to select stable Huh7 cells for another 48 hrs.
Huh-7 cells were transfected with miR-3651 mimics, miR-3651-inhibitor, miR-3651 negative control (miR-NC) for 24 hrs, using lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) following the manufacture’s instruction. In addition, Bel-7402 cells were transfected with miR-3651-inhibitor or miR-NC for 24 hrs using lipofectamine 2000 transfection reagent. MiR‐3651 mimics, miR-3651 inhibitor and miR-NC were purchased from GenePharma (Shanghai, People’s Republic of China).
Real-time reverse transcriptase quantitative PCR (RT-qPCR)
Total RNAs in HCC tissues and Huh-7 cells were isolated using RNAiso Plus (Takara Biotechnology, Co. Ltd. Dalian, People’s Republic of China). Then, complementary DNA was reverse transcribed using a PrimeScript™ RT Master Mix Kit (Takara Biotechnology). Later on, real-time PCR was performed using a SYBR Green™ Premix Ex Taq™ (Takara Biotechnology) with a LightCycler 96 System (Roche Diagnostics Corporation, Indianapolis, IN, USA). U6 and β-actin were used as the internal control of the miR-3651 and PTEN. The primer sequences are as follows: miR-3651 forward: 5’-CCGGTCGCTGGTACATGAC-3’; reverse: 5’-CTCAACTGGTGTCGTGGAGTC-3’, U6 forward: 5’- CTCGCTTCGGCAGCACAT-3’; reverse: 5’-AACGCTTCACGAATTTGCGT-3’, PTEN forward: 5’-ATTCCCAGTCAGAGGCGCTAT-3’; reverse: 5’- GAACTTGTCTTCCCGTCGTGT-3’, β-actin forward: 5’- GTCCACCGCAAATGCTTCTA-3’; reverse: 5’-TGCTGTCACCTTCACCGTTC-3’. Classic 2−ΔΔCt method was used to analyze data.
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was used to determine the cell viability. Huh-7 or Bel-7402 cells (5×103 cells per well) were plated into a 96-well plate and incubated at 37°C overnight, respectively. Then, the cells were transfected with miR-3651 mimics, miR-3651 inhibitor or miR-NC for 24 hrs. After another 48 hrs of incubation, 10 μL of the CCK8 reagent was added to each well for 2 hrs. The OD was detected using an enzyme-linked immunosorbent assay reader at a wavelength of 450 nm.
Immunofluorescence staining
Huh-7 cells were transfected with miR-3651 mimics, miR-3651 inhibitor or miR-NC for 24 hrs and the cells were incubated for another 48 hrs. Then, cells were prefixed in 4% paraformaldehyde for 15 mins, and permeabilized with 100% methanol for 20 mins. After that, cells were stained with primary antibodies at 4°C overnight: anti-Ki67 (Abcam Cambridge, MA, USA; ab15580) (1:1000), DAPI (Abcam; ab104139), followed by goat anti-rabbit IgG H&L (Alexa Fluor® 488) (1:5000, Abcam; ab150077) for 1 hr. Later on, the samples were observed using a Nikon Eclipse TE2000 fluorescent microscope (Japan, Tokyo).
Flow cytometric analysis of cell apoptosis
Cell apoptosis of HCC cells were analyzed by staining with Annexin V-FITC (BD Bioscience, Franklin Lake, NJ, USA) by a flow cytometer. Huh-7 cells were transfected with miR-3651 mimics, miR-3651 inhibitor or miR-NC for 24 hrs and the cells were incubated for another 48 hrs. After that, the cells were collected and washed twice in PBS. Then, the cells were stained with Annexin V-FITC and PI (BD Bioscience) for 30 mins at 37°C in the dark. Apoptotic cells were quantified with a Beckman Coulter (Fullerton, CA, USA) and calculated using FCS Express software (De Novo Software, Los Angeles, CA, USA).
Wound healing assay
Huh-7 cells (4×105 cells per well) were seeded into 6-well plate overnight. At 80% confluent cells, a “scratch” was made in the cell monolayer to create a wound. Then, cells were transfected with miR-3651 mimics or miR-3651 inhibitor for 24 hrs at 37°C. Images were observed at 0 and 24 hrs by fluorescence microscope (Olympus CX23 Tokyo, Japan). The initial wound width (Wi 0 h) and the final wound width (Wf) at 24 hrs after scratching were photographed. Migration distance (Md 24 h)=Wi 0 h – Wf 24 h, wound healing rate (100%)=Md 24 h, miR-3651 mimics/ Md 24 h, NC x 100%.
Transwell migration and invasion assays
Cell migration and invasion assays were performed using 24-well plates inserted with filters (8-mm pore size, Falcon, BD Biosciences). Huh-7 cells were seed into the upper chamber with serum-free medium, and the lower compartment of each well was added with RPMI 1640 containing with 20% FBS. After 24 hrs of incubation, cells on the upper surface were removed. The migrated or invaded cells on the lower surface were fixed with 4% paraformaldehyde and then stained with 0.5% crystal violet. After that, the numbers of migrated or invaded cells in five random fields were visualized (magnification, ×200) and counted. For invasion assay, the upper chambers were pre-treated with 100 μL of Matrigel (BD Bioscience).
Western blotting
Cells were lysed in RIPA Lysis Buffer and a BCA Protein Assay kit (Thermo Fisher Scientific) was used to determine the protein concentration. Proteins of each sample were separated by SDS-PAGE gel. Then, the proteins were transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). After that, the membranes were blocked with 5% non-fat milk, followed by the incubation with primary antibodies at 4°C overnight: anti-Bax (Abcam; ab32503) (1:1000), anti-active caspase 3 (Abcam; ab2302) (1:1000), anti-Bcl-2 (Abcam; ab32124) (1:1000), anti-β-actin (Abcam; ab8227) (1:1000), anti-PTEN (Abcam; ab32199) (1:1000), anti-p-PI3K p85 (p-p85, Abcam; ab182651) (1:1000), anti-p-Akt (Abcam; ab38449) (1:1000). The membranes were washed in TBST three times then incubated with secondary antibodies for 1 hr at room temperature. Chemiluminescence (Millipore Corporation, Billerica, MA, USA) were applied to measure protein expression using densitometry analysis (ImageJ software).
Dual-luciferase reporter assay
Huh-7 cells (4 x 104 cells per well) were seeded into 24-well plates overnight, then wild-type PTEN 3′-UTR (WT-PTEN 3′-UTR) or mutant PTEN 3′-UTR (MT-PTEN 3′-UTR) was co-transfected with miR-3651 mimics or miR-3651 inhibitor, respectively using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, the luciferase activities were detected using a Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). The data were normalized to Renilla luciferase.
Animal study
Five-week-old male nude mice (n=9) was purchased from the Shanghai SLAC Animal Center (Shanghai, People’s Republic of China Animals were divided into three groups (n=3 in the blank group, n=3 in the NC group and n=3 in the miR-3651 inhibitor group). Huh-7 cells (5×106 cells, in 100 μL of PBS) were injected subcutaneously into the right armpit area of each mouse. When the tumors reach 180 mm,3 animals were received an intratumor injection of 50 nM miR-3651 inhibitor or miR-NC twice a week. The volume of the tumors in each group was measured every week following formula: volume (mm3=length×(width2)/2). After 21 days of treatment, the entire tumors were removed from mice and weighted. National Institutes of Health guide for the care and use of laboratory animals was followed in this study. All animal experiments protocol was approved by the Ethics Committees of the Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology.
Statistical analysis
All data were shown as the mean±SD. The data were analyzed with GraphPad Prism version 7 (GraphPad Software, version 7.0, La Jolla, CA, USA). The comparison between two groups was conducted by Student’s t-test. All experiments were performed at least three independent experiments. P<0.05 was considered statistically significant.
Results
Levels of miR-3651 were upregulated in HCC tissues
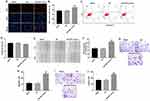
To investigate the role of miR-3651 in HCC tumorigenesis, the levels of miR-3651 in 30 samples of HCC tissues and adjacent normal tissues were assessed using qRT-PCR. As is shown in Figure 1A, high expression of miR-3651 was observed in HCC tissues compared to the adjacent normal tissues. In addition, clinic-pathological parameters displayed that higher miR-3651 level was positively correlated with tumor volume and lymph node metastasis in patients with HCC (Table 1). To investigate the biological role of miR‑3651 in HCC, the level of miR-3651 was significantly increased following transfection with miR-3651 mimics (Figure 1B). In addition, a CCK-8 assay demonstrated that overexpression of miR-3651 markedly promoted the proliferation of Huh-7 cells (Figure 1C). Therefore, these results suggested that miR-3651 might be an oncogene in HCC.
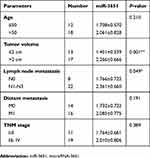 |
Table 1 Level of miR-3651 and clinic-pathological parameters of patients with hepatoma |
Overexpression of miR-3651 promoted the proliferation and migration of HCC cells
Next, Ki67 immunofluorescence assay was performed to validate the proliferation of Huh-7 cells following transfection with miR-3651 mimics. As revealed in Figure 2A and B, overexpression of miR-3651 obviously promoted proliferation ability of Huh-7 cells compared with NC group. In addition, the apoptosis rate of Huh-7 cells was not affected by transfection with miR-3651 mimics (Figure 2C and D). Moreover, wound healing and transwell migration assays were conducted to explore the effect of miR-3651 mimics on HCC cell migration. The results indicated that overexpression of miR-3651 obviously increased migration ability of Huc-7 cells (Figure 2E–H). Meanwhile, overexpression of miR-3651 markedly increased invasion ability of Huc-7 cells (Figure 2I and J). Thus, our results revealed that overexpression of miR-3651 could promote proliferation, migration and invasion in HCC cells.
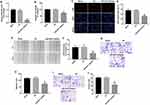
Downregulation of miR-3651 inhibited the proliferation and migration of HCC cells
To further explore the function of miR-3651 in HCC, miR-3651 inhibitor was transfected into Huh-7 cells. QRT-PCR illustrated that the level of miR-3651 in cells was markedly decreased following transfection with miR-3651 inhibitor compared with the NC group (Figure 3A). CCK-8 and immunofluorescence assay displayed that inhibition of miR-3651 significantly suppressed the viability and proliferation of Huh-7 cells, compared with the NC group (Figure 3B–D). Meanwhile, we found that inhibition of miR-3651 obviously suppressed the viability of Bel-7402 cells (Figure S1A and B), which were consistent with the data from the Huh-7 cells. As expected, downregulation of miR-3651 markedly inhibited the ability of migration in Huh-7 cells (Figure 3E–H). Similarly, silencing of miR-3651 markedly attenuated the ability of invasion in Huh-7 cells (Figure 3I and J). These data revealed that downregulation of miR-3651 could inhibit proliferation, migration and invasion in HCC cells.
Downregulation of miR-3651 induced apoptosis in HCC cells
Next, flow cytometric assay was applied to assess the effect of miR-3651 inhibitor on HCC cell apoptosis. As shown in Figure 4A and B, miR-3651 inhibitor significantly induced apoptosis of Huh-7 cells compared with the NC. Similarly, downregulation of miR-3651 obviously induced apoptosis of Bel-7402 cells (Figure S1C and D). In addition, Western blotting assay revealed that downregulation of miR-3651 markedly decreased the level of Bcl-2 and increased the expressions of Bax and active caspase 3 in cells (Figure 4C–F). All these data suggested that downregulation of miR-3651 could induce apoptosis of HCC cells.
PTEN was a direct binding target of miR‑3651
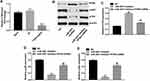
Online bioinformatics tools TargetScan, miRDB, and miRNA were used to search target genes of miR-3651. The data indicated that PTEN might be a potential target of miR-3651 (Figure 5A). In addition, we conducted dual-luciferase reporter assay to confirm whether PTEN was a binding target for miR-3651. The results demonstrated that miR-3651 mimics inhibited the luciferase activity of psiCHECK-2-PTEN-WT, but it did not affect the luciferase activity of psiCHECK-2-PTEN-MT (Figure 5B). In contrast, miR-3651 inhibitor enhanced the luciferase activity of psiCHECK-2-PTEN-WT (Figure 5C). Moreover, qRT-PCR assay showed that the level of PTEN was decreased in Huh-7 cells following transfection with miR-3651 mimics (Figure 5D). Meanwhile, the results of Western blot indicated that the expression of PTEN was markedly decreased, whereas p-p85 and p-Akt were notably increased following transfection with miR-3651 mimics compared with NC group (Figure 5E–H). All these data suggested that PTEN was a direct binding target of miR‑3651.
Downregulation of miR-3651 inhibited growth of HCC cells via upregulating PTEN
To further investigate whether miR-3651 was capable of regulating the growth of HCC cells by targeting PTEN, we downregulated PTEN in Huh-7 cells. As shown in Figure 6A, the level of PTEN was significantly downregulated in Huh-7 cells following infection with PTEN-shRNA. In addition, the PTEN-shRNA and miR-3651 inhibitor plasmids were co-transfected into Huh-7 cells. The results indicated that downregulation of miR-3651 markedly increased the level of PTEN, and decreased the expressions of p-p85 and p-Akt in Huh-7 cells. However, these effects were markedly reversed in Huh-7 cells following infection with PTEN-shRNA, compared with miR-3651 inhibitor group (Figure 6B–E). These data confirmed that downregulation of miR-3651 inhibited the growth of HCC cells via upregulating PTEN.
Downregulation of miR-3651 inhibited HCC tumor growth in vivo
To assess the effect of miR-3651 on HCC tumor growth in vivo, tumor xenograft model was established. As indicated in Figure 7A and B, downregulation of miR-3651 markedly repressed tumor growth of xenograft. In addition, downregulation of miR-3651 significantly reduced the weight of tumors, compared with the NC group (Figure 7C). As shown in Figure 7D, the level of miR-3561 was significantly downregulated in tumor tissues after injection with miR-3651 inhibitor, compared with the NC group. We next explored the effects of miR-3651 on the expression of apoptosis-related proteins in tumor tissues. Western blotting assay indicated that the expression of PTEN was notably increased and the expression of p-Akt was obviously decreased in miR-3651 inhibitor group, compared with the NC group (Figure 7E–G). These data revealed that downregulation of miR-3651 could inhibit HCC cell growth in vivo.
Discussion
Previous studies characterized the miRNA expression profiles related with growth and metastasis and prognosis of HCC, and dysregulation of miRNAs plays a vital role in hepatic carcinogenesis.17,18 miRNAs could act as potential biomarkers in HCC.19 In the previous study, miR-3651 was significantly upregulated in nasopharyngeal carcinoma.20 And it was markedly downregulated in esophageal squamous cell cancer.14 Taken together, miR-3651 might function as a tumor oncogene or suppressor, depending on the tumor types. In this study, we found that the level of miR-3651 was upregulated in HCC tissues. Moreover, high expression level of miR-3651 was associated with the tumor volume and lymph node metastasis. These data preliminary indicated that miR-3651 functioned as an oncogene in HCC.
Evidence revealed that miR-3651 acted as a novel biomarker in HCC.16 However, the mechanisms by which miR-3651 regulates the proliferation, apoptosis and invasion in HCC remain unclear. Therefore, to investigate the function of miR-3651 in HCC, in vitro and in vivo experiments were performed. Our results illustrated that overexpression of miR-3651 significantly promoted cell proliferation and invasion, whereas downregulation of miR-3651 markedly inhibited cell growth and invasion in HCC. In addition, downregulation of miR-3651 could induce apoptosis of Huh-7 cells via upregulating the expressions of pro-apoptotic proteins Bax, active caspase 3 and downregulating the level of anti-apoptotic protein Bcl-2.
It has been indicated that miRNAs exert their functions mainly based on their target genes. To investigate the underlying mechanism of miR-3651 in HCC progression, bioinformatics analysis and luciferase reporter assay were applied. We found PTEN was identified as a direct target of miR-3651 in HCC. PTEN is a tumor suppressor gene, which could negatively regulate the phosphoinositide 3-kinase (PI3K) pathway.21 In addition, PTEN could regulate proliferation, metastasis and apoptosis of HCC.13,22 Moreover, PTEN silencing was positively associated with the migration, invasion and proliferation in HCC.23 Cui et al indicated that upregulating PTEN could deactivate the PI3K/Akt pathway.24 Sun et al revealed that miR-486-5p inhibited apoptosis in cardiomyocytes via inhibiting the expression of PTEN, and activating the PI3K/Akt signaling pathway.25 In this study, upregulation of miR-3651 significantly decreased the level of PTEN, and increased the levels of p-p85 and p-Akt in Huh cells. However, downregulation of miR-3651 significantly increased the level of PTEN, and decreased of p-Akt expression in xenograft model. Our findings were consistent with previous studies. Collectively, our findings revealed that miR-3651 exerted oncogenic effect on Huh-7 cells via inhibiting the level of PTEN, and activating the PI3K/Akt pathway.
Conclusion
This study revealed that high-expressed miR-3651 is an oncogenic miRNA that facilitates the progression of HCC through inhibiting PTEN. Therefore, miR-3651 might be a potential biomarker and therapeutic target for the treatment of HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tao X, Yang X, Wu K, et al. miR-629-5p promotes growth and metastasis of hepatocellular carcinoma by activating beta-catenin. Exp Cell Res. 2019. doi:10.1016/j.yexcr.2019.03.042.
2. Yang L, Peng X, Li Y, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer. 2019;18(1):78. doi:10.1186/s12943-019-1010-6
3. Liu CJ, Kao JH. Global perspective on the natural history of chronic hepatitis B: role of hepatitis B virus genotypes A to J. Semin Liver Dis. 2013;33(2):97–102. doi:10.1055/s-0033-1345716
4. Dong L, Li G, Li Y, Zhu Z. Upregulation of long noncoding RNA GAS5 inhibits lung cancer cell proliferation and metastasis via miR-205/PTEN axis. Med Sci Monit. 2019;25:2311–2319. doi:10.12659/MSM.912581
5. Wei KR, Yu X, Zheng RS, et al. Incidence and mortality of liver cancer in China, 2010. Chin J Cancer. 2014;33(8):388–394. doi:10.5732/cjc.014.10088
6. Kessler SM, Hosseini K, Hussein UK, et al. Hepatocellular carcinoma and nuclear paraspeckles: induction in chemoresistance and prediction for poor survival. Cell Physiol Biochem. 2019;52(4):787–801. doi:10.33594/000000055
7. Jiang J, Chen Y, Dong T, et al. Polydatin inhibits hepatocellular carcinoma via the AKT/STAT3-FOXO1 signaling pathway. Oncol Lett. 2019;17(5):4505–4513. doi:10.3892/ol.2019.10123
8. Tang YH, He GL, Huang SZ, et al. The long noncoding RNA AK002107 negatively modulates miR-140-5p and targets TGFBR1 to induce epithelial-mesenchymal transition in hepatocellular carcinoma. Mol Oncol. 2019. doi:10.1002/1878-0261.12487.
9. Dutta R, Mahato RI. Recent advances in hepatocellular carcinoma therapy. Pharmacol Ther. 2017;173:106–117. doi:10.1016/j.pharmthera.2017.02.010
10. Goeppert B, Truckenmueller F, Ori A, et al. Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p. Sci Rep. 2019;9(1):4796. doi:10.1038/s41598-019-40857-3
11. Macharia LW, Wanjiru CM, Mureithi MW, et al. MicroRNAs, hypoxia and the stem-like state as contributors to cancer aggressiveness. Front Genet. 2019;10:125. doi:10.3389/fgene.2019.00125
12. Dai M, Li L, Qin X. Clinical value of miRNA-122 in the diagnosis and prognosis of various types of cancer. Oncol Lett. 2019;17(4):3919–3929. doi:10.3892/ol.2019.10024
13. He J, Mu M, Luo Y, et al. MicroRNA-20b promotes proliferation of H22 hepatocellular carcinoma cells by targeting PTEN. Oncol Lett. 2019;17(3):2931–2936. doi:10.3892/ol.2019.9925
14. Wang C, Guan S, Chen X, et al. Clinical potential of miR-3651 as a novel prognostic biomarker for esophageal squamous cell cancer. Biochem Biophys Res Commun. 2015;465(1):30–34. doi:10.1016/j.bbrc.2015.07.109
15. Ries J, Vairaktaris E, Agaimy A, et al. miR-186, miR-3651 and miR-494: potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol Rep. 2014;31(3):1429–1436. doi:10.3892/or.2014.2983
16. Zhu HR, Huang RZ, Yu XN, et al. Microarray expression profiling of microRNAs reveals potential biomarkers for hepatocellular carcinoma. Tohoku J Exp Med. 2018;245(2):89–98. doi:10.1620/tjem.245.89
17. Jin W, Zhong N, Wang L, et al. MiR-331-3p inhibition of the Hepatocellular Carcinoma (HCC) Bel-7402 cell line by down-regulation of E2F1. J Nanosci Nanotechnol. 2019;19(9):5476–5482. doi:10.1166/jnn.2019.16535
18. Lin X, Xiaoqin H, Jiayu C, et al. Long noncoding RNA miR143HG, low expression in hepatocellular carcinoma, predicts a good prognosis and inhibits tumor multiplication and metastasis by suppressing MAPK and Wnt signaling pathways. Hepatol Res. 2019. doi:10.1111/hepr.13344.
19. Wen Y, Han J, Chen J, et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137(7):1679–1690. doi:10.1002/ijc.29544
20. Peng J, Feng Y, Rinaldi G, et al. Profiling miRNAs in nasopharyngeal carcinoma FFPE tissue by microarray and next generation sequencing. Genom Data. 2014;2:285–289. doi:10.1016/j.gdata.2014.08.005
21. Bertram J, Peacock JW, Tan C, et al. Inhibition of the phosphatidylinositol 3’-kinase pathway promotes autocrine fas-induced death of phosphatase and tensin homologue-deficient prostate cancer cells. Cancer Res. 2006;66(9):4781–4788. doi:10.1158/0008-5472.CAN-05-3173
22. Wang H, Zhao Y, Chen T, et al. MiR-371 promotes proliferation and metastasis in hepatocellular carcinoma by targeting PTEN. BMB Rep. 2019. doi:10.5483/BMBRep.2019.52.5.155.
23. Han Y, Chen M, Wang A, Fan X. STAT3-induced upregulation of lncRNA CASC11 promotes the cell migration, invasion and epithelial-mesenchymal transition in hepatocellular carcinoma by epigenetically silencing PTEN and activating PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2019;508(2):472–479. doi:10.1016/j.bbrc.2018.11.092
24. Cui C, Li S, Wu D. Znhit1 inhibits breast cancer by up-regulating PTEN to deactivate the PI3K/Akt/mTOR pathway. Life Sci. 2019. doi:10.1016/j.lfs.2019.03.067
25. Sun XH, Wang X, Zhang Y, Hui J. Exosomes of bone-marrow stromal cells inhibit cardiomyocyte apoptosis under ischemic and hypoxic conditions via miR-486-5p targeting the PTEN/PI3K/AKT signaling pathway. Thromb Res. 2019;177:23–32. doi:10.1016/j.thromres.2019.02.002
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.