Back to Journals » Cancer Management and Research » Volume 12
MicroRNA-361-3p Inhibit the Progression of Lymphoma by the Wnt/β-Catenin Signaling Pathway
Authors Zhou H , Tang H , Li N , Chen H, Chen X, Gu L, Zhang L, Tian G, Tao D
Received 9 July 2020
Accepted for publication 5 November 2020
Published 2 December 2020 Volume 2020:12 Pages 12375—12384
DOI https://doi.org/10.2147/CMAR.S270374
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Hui Zhou,1 Huifeng Tang,1 Ning Li,1 Hang Chen,1 Xiaohui Chen,1 Lei Gu,1 Liang Zhang,1 Guoyan Tian,1 Diehong Tao2
1Department of Hematology and Oncology, The Affiliated Hospital of Hangzhou Normal University, Hangzhou 310015, People’s Republic of China; 2Hematology Department, Tongde Hospital, Zhejiang Province, Hangzhou 310003, People’s Republic of China
Correspondence: Hui Zhou
Department of Hematology and Oncology, The Affiliated Hospital of Hangzhou Normal University, 126# Wenzhou Road, Hangzhou 310015, People’s Republic of China
Tel +86-571-88358150
Email [email protected]
Diehong Tao
Hematology Department, Tongde Hospital of Zhejiang Province, #345 Gucui Road, Hangzhou 310003, People’s Republic of China
Tel +86-571-89972417
Email [email protected]
Background: MicroRNA is involved in the development of lymphoma. It is reported that miR-361-3p has a tumor inhibitory effect, but its role in lymphoma is still unclear. The purpose of this study is to examine whether miR-361-3p can inhibit the development of lymphoma and further explore the related potential mechanism.
Methods: In this study, we first analyzed the biological function of miR-361-3p in transfected Raji that mimicked miRNA. We also analyzed the biological function of the whole population in stably expressed miR-361-3p transgenic cells. Next, we conducted a complete micro-gene network to test the genetic profile of differential expression of stable gene-modified cells.
Results: We found that miR-361-3p expression was often reduced in lymphoma cell lines. Cellular assays have shown a significant role in inhibiting the growth of miR-361-3p by inhibiting lymphoma proliferation and migration, and severely inhibiting the Wnt/β-catenin series protein signal. Bioinformatics analysis shows that Wnt10A is a new target of miR-361-3p, which is confirmed by our mechanism research. It is confirmed that restoring Wnt10A can reduce the tumor inhibition of Wnt/β-catenin during lymphoma progression and restore the normal signal of Wnt/β-catenin series proteins.
Discussion: Our data indicate that miR-361-3p inhibits the Wnt/β-catenin protein signal by locking Wnt10A, which is an important factor in inhibiting the tumor in the pathogenesis of lymphoma. The miR-361-3p/Wnt10A axis may be a promising target for the treatment of lymphoma.
Keywords: miRNA, lymphoma, Wnt, pathway, tumor suppressor
The number of lymphoma cases is increasing, which is insensitive to a large number of patients treated by standard therapy. Accurate diagnosis, prediction, and treatment still need other indicators for diagnosis, prediction, and clinical outcome evaluation. The imbalance or function of miRNA expression in various types of lymphoma is related to the pathogenesis of lymphoma. Basically, all lymphomas seem to have unique specific miRNA profiles, and some miRNAs are associated with treatment resistance or have unique kinetic properties during treatment.1 Therefore, they may be specific biomarkers for the diagnosis of lymphoma. It can also be used to evaluate pre-treatment or pathological reactions, especially to evaluate early detection of recurrence, and make a significant contribution to clinical decision-making.
Micronucleic acids (miRNAs) are nucleic acid molecules, which are regulated by short-term non-protein coding (about 22 nucleotides).2 Since the discovery of the first lin-4 microRNA in nematode in 1993,3,4 it is estimated that nearly 60% of mammalian protein coding genes have been regulated by iRNAs, and this number will definitely increase with future research.5 All miRNAs are completely bound to mRNA targets to inhibit gene expression, usually associated with discontinuous 3ʹUTR or rarely attached to discontinuous 5ʹUTR of mRNA. When mRNA is attached to the relevant regions of mRNA, the translation of mRNA is reduced and mRNA is destroyed.6 Some reports describe microRNA as oncogenes or tumor suppressors. However, many microRNA have also been shown to act as oncogenes or tumor suppressor genes in various types of cancers.7–9 MiR-361-3p can inhibit the proliferation and metastasis of tumor cells in some cancers, such as non-small cell lung cancer,10 prostate cancer,11 and cervical cancer.12 However, the role of miR-361-3p in lymphoma is unclear.
Therefore, our experiments were performed to investigate the biological function of miR-361-3p in lymphoma cells. Luciferase assays and other molecular experiments were conducted to elucidate the mechanisms underlying miR-361-3p mediated regulation of lymphoma progression. Our study identified miR-361-3p as a novel therapeutic and predictive target for lymphoma. This research was approved by the Affiliated Hospital of Hangzhou Normal University and was carried out according to the principles outlined in the Helsinki Declaration.
Materials and Methods
Bioinformatic and Statistical Analyses
The expression profile data of GSE31377 gene array was downloaded from the open comprehensive database of gene expression, which included six pairs of normal tissues and 50 lymphoma tissues. This data set was analyzed using GEO2R, and a scatter plot for the expression pattern analysis was obtained.
Cell Lines and Cell Culture
The human lymphoma cell lines Jurkat, Hut102, U937 and Raji were purchased from ATCC (https://www.atcc.org/). The cells were cultured according to the standard protocols. These cell lines and normal human lymphocytes were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum, incubated in a 5% CO2 incubator at 37°C. According to cell demands, the medium was refreshed every day or every other day.
Real-Time PCR with Reverse Transcription
Total cellular nucleic acid was separated by triazole reagent (Invitrogen, USA), dissolved in H2O, and stored at −80°C without nucleic acid. cDNA was then synthesized according to the manufacturer’s plan using SYBR Green qRT-PCR (Thermo Fisher Scientific) kit. Subsequently, reverse transcription polymerase chain reaction based on cDNA was carried out on real-time detectors (ABI, ABI-7300, USA). Quantitative and real-time chain reaction of polymerase was carried out on an ABI 7900 high temperature testing instrument (ABI city, New York, USA).
The real-time PCR was as follows: 1) 95°C for 10 minutes; 2) 95°C for 15 seconds; 3) 60°C for 45 seconds; 4) Repeat steps 2–3 40 times; 5) 95°C for 15 seconds; (6) Heat insulation at 60°C for 1 minute; 7) 95°C for 15 seconds; 8) 60°C for 15 seconds, and 9) 4°C forever. The experiments were carried out in triplicate for each data point.
The sequences of the primer pairs were:
RT-Primer (5ʹ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATCA 3ʹ),
PCR Primer:
forward (5ʹ CGTCCCCCAGGTGTGATTC 3ʹ),
reverse (5ʹAGTGCAGGGTCCGAGGTATT 3ʹ).
The results were analyzed by the reference loop method using the software included with the instrument (ABI Prism 7300 SDS Software).
Cell Transfection
As part of this study, cell transfer was carried out to change the expression level of miR-361-3p. The miR-361-3p mimic and miR-361-3p inhibitor were designed and synthesized by a gene pharmaceutical company in Shanghai, China.
The miRNA mimic sequence is as follows: miR‐361-3p mimic: 5ʹ-UCCCCCAGGUGUGAUUCUGAUUU-3ʹ, miR‐361-3p inhibitor: 5ʹ-AAAUCAGAAUCACACCUGGGGGA-3ʹ.
Cells were inoculated into 6-well plates 24 hours prior to transfection. When the cell fusion reached 60–70%, the cells were transfected with liposome 2000 (Invitrogen) quickly. After staining for 6 hours, the medium was changed, and after being cultured for 48 hours, the cells were collected for subsequent experiments.
Cell Counting Kit-8 Assay
According to the manufacturer’s procedure, cell viability was evaluated by analyzing cell counting kit-8 (CCK8, SAB, CP002). Logarithmic growth phase lymphocytes of each transfection group were inoculated in a 96-well plate with a density of 2×105 cells per well (100 μL per well).
Then, cells were cultured in a CO2 incubator at 37°C for 12, 24, 48, and 72 hours respectively. After transfection, the cells were treated with CCK-8 and cultured in 37°C medium for 1 hour. Next, the absorption value (optical density) of each hole was measured to be 450 nm.
Cell Apoptosis Analysis
The Annexin V-FITC/PI apoptosis detection kit (Beyotime, China) was used to detect apoptosis according to the manufacturer’s procedure, as described above. Cells were harvested with trypsin/EDTA washed with PBS, and then resuspended by adding binding buffer. Then, the cells were incubated with 5 μL of V-FITC membrane-binding protein and 5 μL of polyimide for 10 minutes in the dark at 25°C.
Finally, a CytoFLEX flow cytometer (Beckman, USA) was used to quantify apoptotic cells.
Western Blot Analysis
In order to carry out Western blot analysis, cell lysate was prepared from cell lines by using the buffer solution preparation kit of RIPA (Jrdun Biotechnology Co., Ltd. Shanghai, China), and quantitative protein concentration by using Bio-Rad protein assay (Biological Wheel, Hercules, CA). Total protein sample (25 μg/μg/hydration channel) was separated, and its proportion was 10% or 12% polyethylene glycol sulfate (SDS-PAGE) and transferred to polyvinylidene difluoride membrane (PVDF; EMD Millipore, Billerica, MA, USA). Then, primary antibodies of target proteins, such as Wnt10A (Abcam, Ab106522), CyclinD1 (CST, #2922), Caspase3 (Abcam, Ab2302), Mcl1 (Abcam, Ab32087), or GAPDH (CST, #5174), and β-catenin (CST, #8480), were diluted to 1:2,000–3,000 in TBST. After incubating the primary antibody at 4°C overnight, the membrane was washed 3-times in TBST to remove unbound primary antibody. After that, the membrane was then incubated with a goat anti-rabbit secondary antibody (Biyuntian, A0208, China) for 1 hour at RT.
Finally, the protein domain was visualized by ECL Technion Gaoxin (Shanghai Taineng Technology Co., Ltd., China) and analyzed by Bio-Rad ChemiDocXRS imaging system (Hercules, CA, USA).
Dual-Luciferase Reporter Gene Assay
The relationship between miR-361-3p and Wnt10A is estimated by https://www.ncbi.nlm.nih.gov/nuccore/NM_025216. Untranslated region 3(UTR) sequence of Wnt10A was synthesized and inserted into a pGL3 promoter. The Mutant fraction (Mut) with miR-361-3p mutation binding site was also cloned into Wnt10A 3ʹUTR of pGL3-Promoter. The pGL3-Promoter-wtWnt10A and pGL3-Promoter-mutWnt10A were delivered to Raji cells with miR-361-3p, respectively. Forty-eight hours after transfection, cells were collected and lysed. Then, the luciferase activity was evaluated by using the luciferase activity detection kit (Promega).
Statistical Analysis
Mean and SDs were used to represent all data and error bars. In the cell experiments, two data sets were compared by T test, two elements were compared by one-way comparative analysis, and multiple comparisons were compared by Dunnett’s test. The difference P<0.05 was considered statistically significant.
Results
The Expression of miR-361-3p in Lymphoma Tissues Based on GEO Datasets
To evaluate how miR-361-3p expression in lymphoma tissues compares to its expression level in normal lymphocytes, we analyzed the microarray data of a geostationary orbit data set. It is worth noting that miR-361-3p in lymphoma tissue was much less than that expressed in normal tissues, respectively (Figure 1).
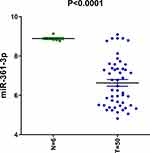 |
Figure 1 miR-361-3p had decreased expression in lymphoma cells. |
The Expression of miR-361-3p on Lymphoma Cell
To determine the effect of miR-361-3p on lymphoma cell, at first, we evaluated the expression level of miR-361-3p in four human lymphoma cells (Jurkat, Hut102, U937, and Raji) and normal lymphocytes. The miR-361-3p expression levels were the lowest in Raji and the highest in normal lymphocytes (Figure 2).
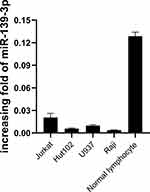 |
Figure 2 qPCR detects the expression of miR-361-3p in the lymphoma cell lines. |
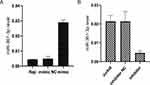
Then, to study the potential function of miR-361-3p in lymphoma, we transferred the MiR-361-3p mimic into Raji cells and the miR-361-3 inhibitor into Jurkat cells. The expression miR-361-3p increased sharply in the miR-361-3p mimic-transfected cells (Figure 3A), while the expression miR-361-3p decreased significantly in the miR-361-3p inhibitor-transfected cells (Figure 3B).
miR-361-3p Promotes Lymphoma Cell Proliferation
The CCK-8 experiment was used to measure the importance of miR-361-3p in lymphoma growth and proliferation. We observed an increase in miR-361-3p expression in Raji cells to inhibit the growth rate of Raji cells, while inhibiting miR-361-3p promoted Jurkat cell proliferation. The difference between them was significant (P<0.05, Figure 4).
 |
Figure 4 miR-361-3p inhibits lymphoma cell proliferation. The effects of miIR-361-3p mimics and inhibitor on Raji (A) and Jurkat (B) cells proliferation as determined by the MTT assay. |
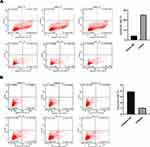
miR-361-3p Promotes the Apoptosis of Lymphoma Cells
In order to further study whether miR-361-3p affects the apoptosis of lymphoma cells, we analyzed Raji and Jurkat cell lines by flow cytometry. We observed a much higher percentage of apoptotic cells in Raji than in normal lymphocytes, with miR-361-3p mimic (25.24%±1.25% vs 4.25±0.06%, P<0.01, Figure 5A), meanwhile the apoptosis rate was significantly lower in Jurkat cells transfected with miR-361-3p inhibitor than in normal lymphocytes (1.08%±0.09% vs 3.89±0.07%, P<0.01, Figure 5B). Thus, these results indicate that miR-361-3p can significantly promote the apoptosis of lymphoma cell lines.
Wnt10A is a Target of miR-361-3p
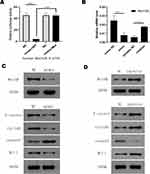
The relationship between miR-361-3p and Wnt10A was determined by bioinformatics prediction and luciferase activity determination. As shown in Table 1, Wnt10A 3ʹ-UTR contained the binding sequences for miR-361-3p. The co-expression of miR-361-3p strongly inhibited the reported activity of fluorescent fluorescence of the wild nature of Wnt10a 3ʹ-UTR, but there was no change in the activity of the Wnt10a 3ʹ-UTR construction block (Figure 6A). The results show that miR-361-3p suppresses the expression Wnt10A by directly combining 3ʹ-UTR.
 |
Table 1 Wnt10A 3ʹ-UTR Contained the Binding Sequences for miR-361-3p |
The real-time quantitative RT-PCR showed that excessive expression of miR-361-3p reduced Wnt10A mRNA and protein levels in Raji cells and that inhibition of miR-361-3p increased Wnt0A expression in Jurkat cells (P<0.001; Figure 6B).
Otherwise, Western blot analysis showed that excessive expression of miR-361-3p lowered the expression of Wnt10A, Cyclin D1, β-catenin, and Mcl-1, and increased the expression of cleaved-caspase3 in Raji cells (Figure 6C); also, inhibition of miR-361-3p upregulated the expression of Wnt10A, Cyclin D1, β-catenin, and Mcl-1, and downregulated cleaved-caspase3 expression in Jurkat cells (Figure 6D).
Taken together, these results demonstrate that miR-361-3p promote lymphoma cell apoptosis by Wnt/β-catenin pathway.
Detection of Wnt10A Expression in Cell Lines
Low expression of miR-361-3p in lymphocyte cells and strong Wnt10A expression (Figure 7).
Discussion
Lymphoma is a group of heterogeneous tumors with different manifestations, prognosis, and mechanisms of morbidity.13 A better understanding of molecular processes in the development and progression of lymphoma can improve early diagnosis and provide more effective therapeutic strategies.
Oligonucleotides are unencrypted short nucleotides of 18–24 nucleotides, which are combined with complementary areas generally located in the target gene 3ʹ UTR (unrecycled zone). They are mainly used as inhibitory molecules, causing post-transcription inhibition or degradation. However, they can also be used as gene activators in some cases.14 According to analysis, microRNA can directly regulate most human genes, accounting for more than 60%.15 Thus, microscopic nucleic acid participates in most, if not all, cellular processes under physiological conditions and can be activated by combining complementary sequences in the target gene. Role of carcinogenic genes or tumor inhibitors.6 In addition, the dysfunctional expression of microscopic nucleic acid appears to be the symbol of all cancers.16,17 As a result, miRNA expression levels can serve as new indicators of diagnosis and may provide new strategies for therapeutic interventions.18 Abnormal miRNA expression in lymphomas (and other cancers) can be caused by many genomic events, such as chromosomal aberrations, visual genetic modifications, sequential mutations of miRNA or its heuristic sub-regions, or adjusting the factors of composition or miRNA function. Abnormal expression of microscopic nucleic acids facilitates diagnosis, prognosis, and prediction of therapeutic responses to lymphomas.19
A previous study showed that miR-361-5p plays a role in tumor inhibition in many cancers.20 It inhibits the development of lung cancer by targeting FOXM1.21 Target CXCR623 to inhibit the growth of liver cancer cells;22 reduce FGFR1 and mmp1 in breast cancer by reducing targeted glycolysis metabolism;23 and by targeting STAT625 to prevent prostate cancer.24 In contrast, there are few studies on the function of miR-361-3p in cancer. Some recent studies have found that miR-361-3p can inhibit the non-small cell lung cancer cell’s proliferation and metastasis by targeting SH2B1 directly.10 Reducing the expression miR-361-3p in prostate cancer could be an important indicator of prostate cancer diagnosis;11 in cervical cancer, the expression miR-361-3p is an independent predictor.12 However, there was no report on the value of miR-361-3p in lymphoma.
Through this study, we can see that the expression level of miR-361-3p in lymphoma cell lines were lower than that in normal tissues and normal lymphocytes. The increased expression of miR-361-3p can reduce the proliferation of lymphoma cells by blocking Wnt10A expression in vitro, and the inhibition of miR-361-3p was enough to cause the up-regulation of Wnt10A expression. These results were in accord with previous reports indicating miR-361-3p as a potential tumor suppressor in human lymphomas.
We also explained the mechanism of proliferation of miR-361-3p lymphoma cells. There was much evidence that the classic Wnt/β-catenin pathways are involved in the appearance and development of tumors, and it is abnormally activated in various malignant hematological diseases. The Wnt signaling pathway is mainly related to hematopoietic activity through its role in the hematopoietic stem cell (HSC) biology,25 which is essential for leukemia development of HSC-derived malignant tumors, and these connections have been extensively studied.26–29 Moreover, Wnt/β-catenin-dependent signaling pathway also plays a key role in malignant hematological diseases derived from mature B cells.30
Data from this study show that Wnt10A was the target of miR-361-3p. First, using the Western blotting, transfection of miR-361-3p mimic has been proved to cause a significant decrease in Wnt10A protein and Wnt/β-catenin pathway genes (including β-catenin, Mcl-1, and CyclinD1) expression (Figure 8). In addition, it was indicated that miR-361-3p was able to directly regulate Wnt10A expression, as the seed region of miR-361-3p is able to bind with the 3ʹ-UTR of Wnt10A mRNA. Furthermore, the luciferase activity of Wnt10a 3ʹ-UTR has a specific reaction to the rise of miR-361-3p. In fact, the mutation in the miR-361-3p binding position eliminated the regulatory effect of the activity of the fluorescent enzyme of miR-361-3p.
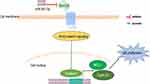 |
Figure 8 A schematic regulation mechanism of miR-361-3p on lymphoma cell behaviors. miR-361-3p inhibits tumor cell proliferation through the Wnt/B-catenin pathway. |
To sum up, our results show that miR-361-3p mediates tumor inhibition in lymphoma, at least partially, ascribed to upregulation of Wnt10A.
In a word, miR-361-3p was down-regulated in lymphoma and regulates the proliferation of lymphoma cells by targeting the Wnt/β-catenin pathway. This microRNA might provide new insights and can be used in the molecular mechanism of lymphoma proliferation and development, and excessive expression of miR-361-3p may be a treatment strategy for lymphoma in the future.
Data Sharing Statement
Please contact the author for data requests.
Consent for Publication
All authors have declared that they agree to publish.
Acknowledgments
This work was supported by grants from the Hangzhou City Health and Family Planning Technology Plan (No. A20200417) and Zhejiang Medical Association Clinical Research Fund Project (No.2018ZYC-A33).
Author Contributions
All authors have made great contributions to the reported work, whether in conception, research design, implementation, data collection, analysis and interpretation, or in all these fields; participated in drafting, modify or comment on articles; give final approval to the forthcoming edition; agreed to submit the article to this periodical; and accept to be responsible for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. De Tullio G, De Fazio V, Sgherza N, etal. Challenges and opportunities of microRNAs in lymphomas. Molecules. 2014;19(9):14723–14781. doi:10.3390/molecules190914723
2. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432(7014):231–235. doi:10.1038/nature03049
3. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi:10.1016/0092-8674(93)90530-4
4. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi:10.1016/0092-8674(93)90529-y
5. Mazan-Mamczarz K, Gartenhaus RB. Role of microRNA deregulation in the pathogenesis of diffuse large B-cell lymphoma (DLBCL). Leukemia Res, 2013;37(11):1420–1428.
6. Tuncer SB, Akdeniz D, Celik B, etal. The expression levels of miRNA-15a and miRNA-16-1 in circulating tumor cells of patients with diffuse large B-cell lymphoma. Mol Biol Rep. 2019;46(1):975–980. doi:10.1007/s11033-018-4554-4
7. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi:10.1016/j.ydbio.2006.08.028
8. He L, Thomson J, Hemann M, etal. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833.
9. Koturbash I, Zemp FJ, Pogribny I, Kovalchuk O. Small molecules with big effects: the role of the microRNAome in cancer and carcinogenesis. Mutat Res Genet Toxicol Environ Mutagen. 2011;722(2):94–105.
10. Chen W, Wang J, Liu S, etal. MicroRNA-361-3p suppresses tumor cell proliferation and metastasis by directly targeting SH2B1 in NSCLC. Journal of Experimental & Clinical Cancer Research,2016;35(1):76.
11. Guzel E, Karatas OF, Semercioz A, etal. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int J Cancer. 2015;136(4):875–9.
12. Liu S, Song L, Yao H, etal. Preserved miR-361-3p expression is an independent prognostic indicator of favorable survival in cervical cancer.Disease markers, 2018;2018:1–9.
13. Swerdlow SH, Campo E, Pileri SA, etal. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi:10.1182/blood-2016-01-643569
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
15. Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008;19(1):92–105. doi:10.1101/gr.082701.108
16. Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133. doi:10.1016/j.gde.2018.01.001
17. Fernandez‐Mercado M, Manterola L, Larrea E, etal. The circulating transcriptome as a source of non‐invasive cancer biomarkers: concepts and controversies of non‐coding and coding RNA in body fluids. Journal of Cellular & Molecular Medicine,2015;19(10):2307–2323.
18. Wang Y, Liang Y, Lu Q. MicroRNA epigenetic alterations: predicting biomarkers and therapeutic targets in human diseases.Clinical Genetics, 2010;74(4):307–315.
19. Solé C, Arnaiz E, Lawrie CH. MicroRNAs as biomarkers of B-cell lymphoma. Biomarker Insights,2018;13:117727191880684.
20. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
21. Hou XW, Sun X, Yu Y, etal. MiR-361-5p suppresses lung cancer cell lines progression by targeting FOXM1. Neoplasma,2017;64(04):526–534.
22. Sun JJ, Chen GY, Xie ZT. MicroRNA-361-5p inhibits cancer cell growth by targeting CXCR6 in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38(2):777–785. doi:10.1159/000443033
23. Ma F, Zhang L, Ma L, etal. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. Journal of Experimental & Clinical Cancer Research,2017;36(1):158.
24. Liu D, Tao T, Xu B, etal. MiR-361-5p acts as a tumor suppressor in prostate cancer by targeting signal transducer and activator of transcription-6(STAT6). Biochemical & Biophysical Research Communications,2014;445(1):151–156.
25. Chen Y, Haviernik P, Bunting KD, Yang YC. Cited2 is required for normal hematopoiesis in the murine fetal liver. Blood,2007;110(8):2889.
26. Gertow K, Hirst CE, Yu QC, etal. WNT3A promotes hematopoietic or mesenchymal differentiation from hESCs depending on the time of exposure. Stem Cell Reports, 2013;1(1):53.
27. Lento W, Congdon K, Voermans C, Kritzik M, Reya T. Wnt signaling in normal and malignant hematopoiesis.Cold Spring Harbor Perspectives in Biology, 2013;5(2).
28. Apple L, Véronique G, Whelan KA, etal. WNT10A promotes an invasive and self-renewing phenotype in esophageal squamous cell carcinoma. Carcinogenesis,2015;5:598–606.
29. Jiang Y, Sheng H, Meng L, Yue H, Liu Y. RBM5 inhibits tumorigenesis of gliomas through inhibition of Wnt/β-catenin signaling and induction of apoptosis.World Journal of Surgical Oncology, 2017;15(1).
30. Janovsk P, Bryja V. Wnt signalling pathways in chronic lymphocytic leukaemia and B‐cell lymphomas. British Journal of Pharmacology, 2017;174(24):4701.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.