Back to Journals » OncoTargets and Therapy » Volume 14
MicroRNA-302s Might Regulate ARL4C-Mediated Gastric Cancer Progression via p53 Signaling: Bioinformatics Analysis and Experiments Validation
Authors Xie N, Pan Y, Wu J, Bai Y, Xiao C, Gao X, Wang J, Liu N
Received 20 September 2020
Accepted for publication 18 January 2021
Published 13 April 2021 Volume 2021:14 Pages 2541—2553
DOI https://doi.org/10.2147/OTT.S282992
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Ning Xie,1,2,* Yifei Pan,3,* Jian Wu,4 Yunfan Bai,3 Cailan Xiao,1,2 Xiaoliang Gao,4 Jinhai Wang,1,2 Na Liu1,2
1Department of Gastroenterology, The Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 2Shaanxi Key Laboratory of Gastrointestinal Motility Disorders, Xi’an Jiaotong University, Xi’an, Shaanxi, People’s Republic of China; 3Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa, Japan; 4Xijing Hospital of Digestive Diseases, Xijing Hospital, The Fourth Military Medical University, Xi’an, Shaanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Na Liu; Jinhai Wang Email [email protected]; [email protected]
Background: Our previous studies demonstrate that ARL4C is the most critical clinical biomarker for gastric cancer (GC) patients among ARL family members (ARLs) and functions as an oncogene in GC. However, its underlying mechanisms in GC need to be further illustrated. In this study, we aim to explore the upstream and downstream molecular mechanisms of ARL4C in GC cells.
Methods: The genetic alteration of ARL4C in GC is analyzed by cBioPortal database. Potential ARL4C-targeted microRNAs (miRs) are predicted by three databases. The high-throughput RNA sequencing is performed to explore the underlying mechanisms of ARL4C in GC cells. The effects of predicted microRNAs on ARL4C, the RNA-sequencing results validation and the biological functions of ARL4C in GC cells are illustrated by in vitro experiments.
Results: Genetic analyses indicate that ARL4C is significantly upregulated in GC, which is not caused by gene amplification. MicroRNAs prediction shows the high relevance between ARL4C and miR-302 members. Moreover, miR-302c or miR-302d transfection reduces ARL4C protein expression in GC cells. Based on the high-throughput RNA sequencing of ARL4C-knockdown cells, enrichment analyses demonstrate that ARL4C is closely related to cell growth and involved in p53 signaling. Moreover, there are strong gene–gene interactions between ARL4C and genes in p53 signaling, and ARL4C downregulation could inhibit the protein expression of MDM2, a critical gene in p53 pathway. Further functional experiments demonstrate that ARL4C silencing leads to cell cycle arrest and increased cell apoptosis in AGS and MKN45 cells.
Conclusion: Our data suggest that miR-302c and miR-302d may function as the upstream regulators of ARL4C. And, ARL4C might promote GC cell cycle progression via regulating p53 signaling. Our findings provide novel insights into the key role of ARL4C and the underlying mechanisms in GC progression, thus facilitating the development of ARL4C-targeted therapy.
Keywords: ARL4C, gastric cancer, miRNA-302s, p53, cell cycle, apoptosis
Introduction
Gastric cancer (GC) is one of the most common malignancies and a main contributor to cancer-related deaths worldwide.1,2 Despite important advances in the diagnosis and treatment for GC, the long-term survival of GC patients is still unsatisfactory.3 Thus, it is imperative to further explore the specific molecular mechanisms underlying the tumorigenesis and progression of GC to pave the way for discovering novel biomarkers with higher specificity and sensitivity for GC.4
The ADP-ribosylation factor-like (ARL) family, as a subfamily of small G proteins, has been implicated in a wide variety of essential cellular functions including vesicular trafficking, cytoskeletal organization, ciliogenesis, cell proliferation, motility and differentiation.5–7 In the recent years, accumulating evidences demonstrate the crucial effects of ARL family members (ARLs) on cancer progression. For instance, ARL5A is reported to be overexpressed in colorectal cancer and its downregulation inhibits the proliferation of colorectal cancer cells.8 ARL13B can promote the progression of GC in vitro and in vivo by regulating Smoothened trafficking and activating Hedgehog signaling.9 ARL11 is established as a tumor suppressor in a variety of solid tumors.10–12 A recent study identifies ARL14 as a crucial prognostic factor of lung adenocarcinoma based on The Cancer Genome Atlas (TCGA) database.13 Furthermore, they discover that downregulation of ARL14 inhibits cell proliferation, migration and invasion as well as attenuates radiation damage of lung adenocarcinoma cells.13 These evidences reveal that targeting ARLs may be a promising precision treatment for cancer. Therefore, the clinical values and biological functions of ARLs in GC arouse our attention.
In our previous study, we demonstrate that ARLs are significantly dysregulated in GC and involved in various cancer-related pathways. Multiple machine learning models identify ARL4C as one of the most important diagnostic and prognostic indicators among ARLs for GC. Moreover, higher ARL4C expression is found in primary GC samples compared with normal gastric mucosa tissues by immunohistochemistry and Western blot. Further experiments demonstrate that down-regulation of ARL4C dramatically inhibits tumorigenesis and metastasis of GC cells in vivo and in vitro. However, the specific molecular mechanisms underlying the tumor-promoting effects of ARL4C on GC cells have not been fully elucidated. Thus, in this study, we aim to further investigate the upstream regulators of ARL4C and the corresponding downstream effectors in GC cells.
Methods
cBioPortal Database Analysis
The cBio Cancer Genomics Portal (cBioPortal) (http://www.cbioportal.org/) provides visualization tools for more than 5000 tumor samples from 232 cancer studies in TCGA database.14,15 In this study, the Stomach Adenocarcinoma (TCGA, Firehose Legacy, n=393) cohort was analyzed. All analyses were performed with default software parameters.
GEPIA Database Analysis
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) is a user-friendly website resource to analyze expression status and prognostic values of specific genes in TCGA.16 In this study, we accessed the Spearman correlation between ARL4C and MDM2 using GEPIA.
Targeted-miRNAs Prediction
RNAInter17 (http://www.rna-society.org/raid/), TargetScan18 (http://www.targetscan.org/vert_72/) and miRDB19 (http://www.mirdb.org/) were utilized to predict the targeted miRNAs of ARL4C.
Metascape Analysis
Metascape (http://metascape.org) is a user-friendly analysis tool for gene annotation and enrichment analysis.20 In this study, Metascape was used to perform Gene Ontology (GO) enrichment analysis of differential genes from high-throughput RNA sequencing and the interaction networks of enrichment terms. All analyses were performed with default software parameters.
GeneMANIA Analysis
GeneMANIA (http://www.genemania.org) is a well-maintained and flexible interactive web server for deriving hypotheses based on gene functions.21 GeneMANIA was adopted to construct a gene–gene interaction network for ARL4C and genes in p53 signaling in this study.
Kaplan–Meier Plotter Analysis
Kaplan–Meier plotter (http://kmplot.com/analysis/) was utilized to analyze the correlation between ARL4C expression and overall survival rates in different GC subtypes based on the hazard ratios (HR) and log-rank P-values.22
Cell Lines
The human gastric carcinoma cell lines (AGS, MKN45) were obtained from the National Infrastructure of Cell Line Resource (China). They were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. All cell lines were cultured at 37°C in a humidified atmosphere containing 5% CO2. Additionally, MKN45 cells had mutated p53.23
Lentivirus Transfection
The human shARL4C (NM_001282431) lentivirus was designed and constructed by GeneChem (Shanghai, China). The negative control (NC) is a scramble shRNA (TTCTCCGAACGTGTCACGT). shARL4C: TCAAGTTCAACGAGTTCGT.
Clinical Samples
Six paired samples of primary GC tissues and adjacent normal tissues were acquired from patients who had undergone GC surgery at Xijing Hospital of Digestive Diseases. All samples were clinically and pathologically verified.
Western Blot Analysis
Cell lysates were prepared using RIPA buffer (HEART, China) including fresh protease and phosphatase inhibitors. The cells were incubated on ice for 30 minutes before centrifugation at 12,000 rpm at 4°C for 15 minutes. Protein concentration was determined using a BCA protein assay kit (HEART, China). The protein samples were boiled in 5 × SDS protein sample buffer (Beyotime, China) for 15 minutes for complete denaturation. Protein (20–50 µg) was separated by SDS-PAGE and transferred to NC membranes (Millipore, USA). Then, the NC membranes were blocked with 5% defatted milk at room temperature (RT) for 2 hours and incubated with primary antibodies at 4°C overnight. The primary antibodies used in our study included anti-ARL4C (at 1:500, ab122025, Abcam, USA), anti-p53 (at 1:1000, #2527, CST, USA), anti-MDM2 (at 1:500, sc-5304, Santa Cruz, USA) and anti-GAPDH (at 1:5000, 10494-1-AP, Proteintech Technology, UK). The membranes were washed with TBS with Tween® 20 (TBST) three times for 5 minutes each, followed by incubation with the corresponding secondary antibodies (MYBiotech, China) at a 1:10,000 dilution for an hour at RT. The membranes were washed as before and visualized using an enhanced chemiluminescent substrate (Millipore, USA).
Luciferase Reporter Assay
Cells were cotransfected with an ARL4C promoter-luciferase reporter plasmid and Renilla luciferase control vector. A Dual-Luciferase Reporter Assay Kit (Promega, USA) was used according to the manufacturer’s instructions to measure luciferase activity after 72 h, and activity was normalized to Renilla luciferase activity. The synthetic sequences were provided by Shanghai Genechem.
RNA Extraction and RNA-Sequencing (RNA-Seq)
Total RNA was extracted from 2 cell lines, AGS-NC and AGS-shARL4C, using TRIzol® Reagent (Invitrogen, USA) according to the manufacturer’s instructions. RNA sequencing was entrusted to Novel Bioinformatics (Shanghai, China). Using high-throughput Life technologies Ion Proton Sequencer, the transcripts with poly(A)-containing human RNA were analyzed. Three biological replicates were performed for each cell. Student’s t-test was used to determine whether the differences of the mean log2(FPKM + 1) values between groups were significant. P < 0.05 was considered statistically significant.
Cell Cycle Analysis and Apoptosis Assays
Single cell suspensions were collected and washed twice with cold PBS. For apoptosis assays, AGS and MKN45 cells were double stained with propidium iodide (PI; Beyotime, Shanghai, China) and Annexin V-fluorescein isothiocyanate (FITC) by an apoptosis detection kit (BestBio, Shanghai, China).
For the cell cycle assay, the cells were fixed with cold 75% ethanol and stored at −20°C for at least 24 h. Then, cells were washed twice with cold PBS before 1 × 106 cells were resuspended in 1 mL PI/RNase staining solution (BD Biosciences, USA) and finally analyzed by a FACSCalibur flow cytometer (BD Biosciences, USA).
Statistical Analysis
Each experiment was carried out at least three times. Statistical analyses were conducted using SPSS statistical software (Version 21.0). All Western blot images were quantified by ImageJ. Images were obtained using GraphPad Prism software (Version 8.0). The counting data were represented by frequency or percentage, and the measurement data were expressed as the mean ± standard deviation (SD). The differences between the two groups were assessed using Student’s t-test. P < 0.05 was considered statistically significant.
Results
The Upregulation of ARL4C May Not Result from the Genetic Alterations
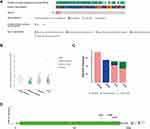
To determine whether the upregulation of ARL4C in GC was caused by genetic alterations, we analyzed the genetic alteration profiles of ARL4C by cBioPortal. Gene alterations of ARL4C were found in 6% of the sequenced samples based on the data obtained from TCGA dataset in cBioPortal (Figure 1A). As shown in Figure 1B, ARL4C mRNA expression was correlated with several copy number changes, including gain, diploid, deep deletion and shallow deletion. Among the cases of diffuse type stomach adenocarcinoma (DTSTAD), stomach adenocarcinoma (STAD) and tubular stomach adenocarcinoma (TSTAD) with gene alterations of ARL4C, mRNA upregulation and deep deletions were the major alteration types. Deep deletions mainly occurred in the cases of mucinous stomach adenocarcinoma (MSTAD) and STAD. While mutations were mostly found in STAD and TSTAD (Figure 1C). Additionally, there were generally three missense mutations (A145V, G165C, A162T) in ARL4C. Detailed changes in protein structure of ARL4C were shown in Figure 1D.
In conclusion, ARL4C was always found overexpressed in GC. However, we found that its high mRNA expression did not result from gene amplification, indicating that alternative mechanism contributed to ARL4C’ upregulation.
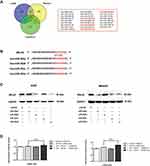
The miR-302 Members May Be Involved in ARL4C-Mediated GC Malignant Progression
By bioinformatics analysis, we screened 26 microRNAs (miRNAs) that might regulate ARL4C in Homo sapiens by RNAInter, TargetScan and miRDB (Figure 2A). Four miR-302 members (miR-302s), including miR-302a, miR-302b, miR-302c and miR-302d, were noticed as they had multiple putative targets at the ARL4C 3ʹUTR (Figure 2B). As is shown in Figure 2C and Supplementary Figure 1A, Western blot analysis indicated that miR-302c and miR-302d transfection could significantly reduce ARL4C protein expression both in AGS and MKN45 cells. Moreover, luciferase assays were performed to validate whether the direct binding affinities between the ARL4C 3′UTR and these two miRNAs existed in HEK 293T cells. Unfortunately, the results showed that ectopic expression of miR-302c or miR-302d could not significantly inhibit the luciferase activity of the constructs that contained the binding sites (Figure 2D). These results revealed that miR-302c and miR-302d could affect the expression of ARL4C possibly by alternative mechanisms, instead of directly binding to ARL4C 3′UTRs.
The Regulating Effects of ARL4C on P53 Signaling May Explain Its Oncogenic Role in GC Cells
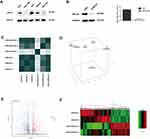
High-throughput RNA-sequencing was carried out to specify the genes that were regulated mainly by ARL4C in GC cells. As shown in Figure 3A and Supplementary Figure 1B, protein expression of ARL4C in different GC cell lines was evaluated by Western blot, and ARL4C was overexpressed in AGS and MKN45 cells. Then, AGS cells were transfected with shRNAs against ARL4C. Multiple clones with stable lentivirus transfection were selected and confirmed by RT-PCR and Western blot analyses (Figure 3B and Supplementary Figure 1C). The six samples were screened for differences in gene expression with a 3:3 ratio. As shown in Figure 3C and D, 2 discrete samples (AGS-NC-1 and AGS-shARL4C-1) were deleted according to the Pearson correlation analysis and principal component analysis (PCA). A total of 641 differentially expressed genes between AGS-shARL4C and AGS-NC groups (FDR < 0.05) had been identified by RNA-sequencing (Figure 3E and F).
Gene Ontology (GO) analysis was used to determine the functions of these differentially expressed genes in Metascape (Figure 4 and Table 1). The top 20 GO enrichment items were classified into three functional groups: biological process (BP) group (15 items), molecular function (MF) group (1 item), and cellular component (CC) group (4 items). The ARL4C-related genes were mainly enriched in several morphogenesis and apoptotic BPs such as negative regulation of apoptotic signaling pathway, response to wounding, positive regulation of apoptotic process involved in morphogenesis, positive regulation of animal organ, vascular wound healing, and reproductive structure development. The MF for these genes was mainly integrin binding. The CCs for these genes were the vacuole, vacuolar lumen, basolateral plasma membrane and collagen-containing extracellular matrix.
 |
Table 1 GO Enrichment Terms of ARL4C-Related Genes in GC Cells |
As shown in Figure 5A and B, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene set enrichment analysis (GSEA) were also performed. The results indicated that p53 signaling pathway had the highest enrichment score for genes co-expressed with ARL4C in AGS. Western-blot analysis showed that the protein expression of ARL4C was significantly negatively correlated with p53 in GC tissues and adjacent normal tissues (Figure 5C and D, Supplementary Figure 1D and 1E). Meanwhile, a gene–gene interaction network constructed by GeneMANIA database also demonstrated that there were multiple interactions between ARL4C and genes involved in p53 signaling, such as co-expression, co-localization, genetic interaction and shared protein domains (Supplementary Figure 2C). By correlation analysis, we found that ARL4C was closely related to MDM2 (Figure 5E), a critical gene in p53 signaling. Meanwhile, ARL4C downregulation could significantly dampen the protein expression of MDM2 in GC cells (Figure 5F, Supplementary Figure 1F and 1G).
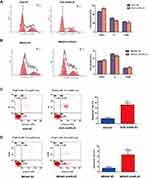
Additionally, we also explored the possible molecular functions of ARL4C in GC by TCGA STAD dataset. The volcano plot in Supplementary Figure 2A showed all ARL4C-associated genes in GC. As shown in Supplementary Figure 2B, KEGG enrichment analysis also demonstrated that ARL4C-related genes were significantly correlated with p53 signaling, cell cycle progression and apoptosis. Meanwhile, further functional experiments also showed that knockdown of ARL4C obviously inhibited cell cycle progression (Figure 6A and B) and elevated cell apoptosis rates (Figure 6C and D) both in AGS and MKN45 cells.
Moreover, we explored the effects of ARL4C on overall survival (OS) of patients with different GC histopathologic subtypes using the Kaplan–Meier (K-M) plotter. As shown in Figure 7A and B, the analysis revealed that higher ARL4C expression was significantly associated with worse OS in intestinal GC which was identified as the p53-mutant molecular subtype.
In a word, our results indicate that ARL4C might regulate the p53 signaling pathway, thus exerting the tumor-promoting effects on GC.
Discussion
Considering that GC is one of the life-threatening malignancies, elucidating the specific mechanisms underlying the development and progression of GC is a priority for researchers. In our previous study, ARL4C has been identified as one of the most important clinical biomarkers for GC patients among ARLs by integrative machine learning models and proved to promote the growth and metastasis of GC cells both in vitro and in vivo. Nevertheless, the upstream and downstream molecular mechanisms of ARL4C in GC cells need to be further illustrated. Our present study aims to provide a more comprehensive understanding of the molecular mechanisms of ARL4C in GC by bioinformatics analysis, high-throughput RNA sequencing and experiments validation.
Our analysis indicates that ARL4C mRNA is overexpressed in GC, but its upregulation is not related to gene amplification. Hence, miRNAs’ regulation on ARL4C is further investigated. We screen out miR-302s as the promising miRNAs which might target ARL4C in GC. miR-302s play various biological roles in diverse cancers by targeting specific genes.24 Moreover, they could participate in regulating several hallmarks of cancer, such as proliferation, cell death and senescence, immune response, angiogenesis, invasion and metastasis.25–27 More importantly, GC patients with decreased expression of miR-302a, miR-302b and miR-302c have aggressive cancer progression and unfavorable clinical outcomes.28,29 Our present experiment shows that miR-302c and miR-302d transfection could significantly decrease the protein expression of ARL4C in GC cells which indicates miR-302c and miR-302d could work as the upstream regulators of ARL4C in GC. Nevertheless, further luciferase assays indicate that neither miR-302c nor miR-302d directly bind to the ARL4C mRNA. Previous studies demonstrate that miR-302s could act as upstream modulators of TGF-β1 signaling members.30 For example, they might directly inhibit the expression of the transforming growth factor beta receptor 2 (TGFBR2) to promote EMT process.31 Our previous study discovers that ARL4C is involved in TGF-β1 signaling and functions as a mediator between TGF-β1 and Smads. Hence, we acknowledge that miR-302c and miR-302d might directly regulate the upstream TGF-β1 signaling molecules of ARL4C to further affect the expression status of ARL4C and ARL4C-mediated GC malignant phenotypes.
In addition, we analyze the changes of genes’ expression in GC cells after ARL4C knockdown using high-throughput RNA sequencing. GO enrichment analysis indicates that ARL4C is highly associated with several morphogenesis and apoptotic processes in GC cells. Further pathway analysis also shows that ARL4C is significantly involved in p53 signaling and interacts with several genes in p53 signaling. The tumor suppressor TP53 is the most frequently mutated gene in GC and is involved in several cell biological processes, such as cell cycle arrest, DNA repair and apoptosis.32,33 Accordingly, we find that ARL4C is highly associated with MDM2 in TCGA database, and ARL4C silencing might decrease MDM2 expression in GC cells. MDM2 is a critical factor in p53 signaling and proved to regulate cell cycle progression, apoptosis and cell growth in GC.34,35 In this study, our functional experiments also illustrate that ARL4C silencing could lead to cell cycle arrest and accelerate cell apoptosis of GC cells. Taken together, our result indicates that ARL4C may play a critical role in GC cell growth via modulating p53 signaling.
On the other hand, GC is mainly characterized into two histopathologic subtypes, intestinal and diffuse GC.36 In addition, GC can be classified into four molecular subtypes based on molecular signatures: EBV-positive (EBV), microsatellite instability (MSI), genomically stable (GS) and chromosomal instability (CIN) according to TCGA.37 Among them, TP53 mutations always occur in the chromosomal instability (CIN) GC, which is mainly the intestinal histological type. Noteworthily, our prognostic analysis also shows that ARL4C is a more significantly negative prognostic factor for patients with intestinal type GC (TP53-mutated) compared to other histological types.
Conclusion
In conclusion, our work is of significance for comprehensively understanding the upstream and downstream molecular mechanisms of ARL4C in GC. Our results demonstrate that miR-302c and miR-302d could function as upstream modulators of ARL4C, and ARL4C could regulate p53 signaling in GC cells. More importantly, we uncover the great promise of ARL4C-targeted therapy in TP53-mutant GC patients. Our studies provide more comprehensive insights into the specific role of ARL4C that benefits the development of novel strategies for GC detection and treatment.
Abbreviations
ARLs, ADP-ribosylation factor-like family members; GC, gastric cancer; TCGA, The Cancer Genome Atlas; cBioPortal, The cBio Cancer Genomics Portal; GO, Gene Ontology; RNA-Seq, RNA-Sequencing; DTSTAD, diffuse type stomach adenocarcinoma; STAD, stomach adenocarcinoma; TSTAD, tubular stomach adenocarcinoma; MSTAD, mucinous stomach adenocarcinoma; miRNAs, microRNAs; miR-302s, miR-302 members; BP, biological process; MF, molecular function; CC, cellular component; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene set enrichment analysis; OS, overall survival; K-M, Kaplan–Meier.
Acknowledgment
The study was supported by grants from the National Natural Science Foundation of China (No. 81872397).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. doi:10.1016/S0140-6736(16)30354-3
3. Dicken BJ, Bigam DL, Cass C, Mackey JR, Joy AA, Hamilton SM. Gastric adenocarcinoma: review and considerations for future directions. Ann Surg. 2005;241(1):27–39. doi:10.1097/01.sla.0000149300.28588.23
4. Vedeld HM, Goel A, Lind GE. Epigenetic biomarkers in gastrointestinal cancers: the current state and clinical perspectives. Semin Cancer Biol. 2018;51:36–49. doi:10.1016/j.semcancer.2017.12.004
5. Burd CG, Strochlic TI, Setty SR. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 2004;14(12):687–694. doi:10.1016/j.tcb.2004.10.004
6. Casalou C, Ferreira A, Barral DC. The role of ARF family proteins and their regulators and effectors in cancer progression: a therapeutic perspective. Front Cell Dev Biol. 2020;8:217. doi:10.3389/fcell.2020.00217
7. Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12(6):362–375. doi:10.1038/nrm3117
8. Wang Q, Huang Z, Guo W. microRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Clin Cancer Res. 2014;20(5):1146–1157. doi:10.1158/1078-0432.CCR-13-1023
9. Shao J, Xu L, Chen L, et al. Arl13b promotes gastric tumorigenesis by regulating Smo trafficking and activation of the hedgehog signaling pathway. Cancer Res. 2017;77(15):4000–4013. doi:10.1158/0008-5472.CAN-16-2461
10. Hamadou WS, Besbes S, Mani R, et al. ARLTS1, potential candidate gene in familial aggregation of hematological malignancies. Bull Cancer. 2017;104(2):123–127. doi:10.1016/j.bulcan.2016.10.016
11. Masojc B, Medrek K, Debniak T, et al. ARLTS1 Trp149Stop mutation and the risk of ovarian cancer. Cancer Res. 2007;67(9):4533. author reply 4534. doi:10.1158/0008-5472.CAN-06-4193
12. Frank B, Hemminki K, Brenner H, Hoffmeister M, Chang-Claude J, Burwinkel B. ARLTS1 variants and risk of colorectal cancer. Cancer Lett. 2006;244(2):172–175. doi:10.1016/j.canlet.2005.12.006
13. Guo F, Yuan D, Zhang J. ARL14Silencing of gene induces lung adenocarcinoma cells to a dormant state. Front Cell Dev Biol. 2019;7:238. doi:10.3389/fcell.2019.00238
14. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
15. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
16. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi:10.1093/nar/gkx247
17. Lin Y, Liu T, Cui T, et al. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020;48(D1):D189–D197. doi:10.1093/nar/gkz804
18. Agarwal V, Bell G, Nam J, Bartel D. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi:10.7554/eLife.05005
19. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(D1):D146–152. doi:10.1093/nar/gku1104
20. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
21. Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: fast gene network construction and function prediction for cytoscape. F1000Research. 2014;3:153. doi:10.12688/f1000research.4572.1
22. Szász A, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget. 2016;7(31):49322–49333. doi:10.18632/oncotarget.10337
23. Naito S, Inoue S, Kinjo M, Tanaka K. Thromboplastic and fibrinolytic activities of cultured human gastric cancer cell lines. Gan. 1983;74(2):240–247.
24. Subramanyam D, Lamouille S, Judson R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29(5):443–448. doi:10.1038/nbt.1862
25. Zhang M, Yang Q, Zhang L, et al. miR-302b is a potential molecular marker of esophageal squamous cell carcinoma and functions as a tumor suppressor by targeting ErbB4. J Exp Clin Cancer Res. 2014;33(1):10. doi:10.1186/1756-9966-33-10
26. Peng N, Yang X, Zhu C, et al. MicroRNA-302 cluster downregulates enterovirus 71-induced innate immune response by targeting KPNA2. J Immunol. 2018;201(1):145–156. doi:10.4049/jimmunol.1701692
27. Maadi H, Moshtaghian A, Taha M, et al. Multimodal tumor suppression by miR-302 cluster in melanoma and colon cancer. Int J Biochem Cell Biol. 2016;81:121–132. doi:10.1016/j.biocel.2016.11.004
28. Ma G, Li Q, Dai W, Yang X, Sang A. Prognostic implications of miR-302a/b/c/d in human gastric cancer. Pathol Oncol Res. 2017;23(4):899–905. doi:10.1007/s12253-017-0282-7
29. Chen L, Min L, Wang X, et al. Loss of RACK1 promotes metastasis of gastric cancer by inducing a miR-302c/IL8 signaling loop. Cancer Res. 2015;75(18):3832–3841. doi:10.1158/0008-5472.CAN-14-3690
30. Balzano F, Cruciani S, Basoli V, et al. MiR200 and miR302: two big families influencing stem cell behavior. Molecules. 2018;23(2):282. doi:10.3390/molecules23020282
31. Gao Z, Zhu X, Dou Y. The miR-302/367 cluster: a comprehensive update on its evolution and functions. Open Biol. 2015;5(12):150138. doi:10.1098/rsob.150138
32. Levine A. p53: 800 million years of evolution and 40 years of discovery. Nat Rev Cancer. 2020;20(8):471–480. doi:10.1038/s41568-020-0262-1
33. Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20(4):199–210. doi:10.1038/s41580-019-0110-x
34. Uchi R, Kogo R, Kawahara K, et al. PICT1 regulates TP53 via RPL11 and is involved in gastric cancer progression. Br J Cancer. 2013;109(8):2199–2206. doi:10.1038/bjc.2013.561
35. Lv B, Ma R, Chen X, et al. E2F1-activated SPIN1 promotes tumor growth via a MDM2-p21-E2F1 feedback loop in gastric cancer. Mol Oncol. 2020;14(10):2629–2645. doi:10.1002/1878-0261.12778
36. Chia N, Tan P. Molecular classification of gastric cancer. Ann Oncol. 2016;27(5):763–769. doi:10.1093/annonc/mdw040
37. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi:10.1038/nature13480
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.