Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-300 suppresses metastasis of oral squamous cell carcinoma by inhibiting epithelial-to-mesenchymal transition
Authors Kang Y , Zhang Y, Sun Y , Wen Y, Sun F
Received 6 May 2018
Accepted for publication 2 July 2018
Published 10 September 2018 Volume 2018:11 Pages 5657—5666
DOI https://doi.org/10.2147/OTT.S173236
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Yuanyuan Kang, Ying Zhang, Yan Sun, Yan Wen, Fuli Sun
Department of Emergency and Oral Medicine, School of Stomatology, China Medical University, Shenyang 110002, China
Introduction: Oral cancer is the sixth most prevalent form of cancer, with oral squamous cell carcinomas (OSCCs) exhibiting the highest morbidity of all head and neck cancers. However, the specific metastatic mechanism is not yet clear. MicroR-300 (miR-300) has been identified as a critical regulator in tumor development. In this study, we focused on the roles of miR-300 in regulating epithelial–mesenchymal transition (EMT) in OSCC.
Methods: The surgical specimens from cases of OSCC (N=120) were collected, and the miR-300 expression was tested and the results were analyzed for possible correlations with clinical characteristics, and the prognostic significance was assessed by Kaplan–Meier analysis and Cox proportional hazards regression in OSCC patients. In addition, the proliferation and invasion of OSCC cells were evaluated after transfection of miR-300 mimics or inhibitor. At last, the role of miR-300 in EMT was investigated.
Results: The results showed that miR-300 levels were significantly lower in patients with OSCC than in controls and miR-300 levels in patients with OSCC were significantly associated with TNM classification. Kaplan–Meier analysis indicated that miR-300 with lower level had significantly decreased overall survival and disease-free survival of OSCC patients. In multivariate analysis, TNM stage, miR-300 expression and tobacco usage were the independent prognostic factors for overall survival and disease-free survival in OSCC. Moreover, in vitro experiments showed that miR-300 could inhibit the proliferation and invasion of OSCC cells. More importantly, miR-300 could inhibit the EMT process. In addition, we found that ET-1 could inhibit the expression of miR-300.
Conclusion: Our findings indicated that miR-300 could suppress metastasis of OSCC by inhibiting EMT. The present study indicates that miR-300 is a potential therapeutic agent for OSCC.
Keywords: OSCC, proliferation, invasion, suppresses, miR-300
Introduction
Oral cancer is the sixth most prevalent form of cancer, with oral squamous cell carcinomas (OSCCs) exhibiting the highest morbidity of all head and neck cancers.1 Although great progress has been made in systemic and individualized treatments of OSCC, the incidence of OSCC has increased significantly over the past few decades2 and metastasis remains a great challenge for the treatment of OSCC. Therefore, it is of great significance to investigate the mechanism of OSCC progression and metastasis.
miRNAs are a group of noncoding RNAs with a length of ~14–22 nucleotides which are expressed in mammalian cells.3,4 It has been found that miRNAs play an important role in various pathological and biological processes such as cell proliferation, migration, differentiation, apoptosis, and inflammation.5–7 Recently, some researchers found that miRNAs can not only regulate the expression levels of oncogenes and tumor suppressor genes to influence the process of tumor, but also might function as oncogenes or tumor suppressor genes directly.8
MiR-300 has been investigated in some human cancers. In gastric cancer, miR-300 was found to be upregulated and overexpression of miR-300 could promote cell proliferation and cell cycle progression.9 In laryngeal squamous cell carcinoma, miR-300 was found to suppress proliferation and invasion of cancer cells by targeting ROS1 and lower miR-300 expression predicts poor prognosis.10,11 It was also found that miR-300 was upregulated in breast cancer and promoted cell proliferation and cell cycle progression by targeting p53.12 This indicated that miR-300 had different functional roles in human cancers. The expression and biological roles of miR-300 in OSCC have not been discovered.
In this study, we report that miR-300 was downregulated in OSCC tissues compared with normal tissues. Decreased expression of miR-300 was associated with TNM classification and unfavorable prognosis of OSCC patients. Furthermore, miR-300 inhibited the proliferation and invasion of OSCC cells in vitro. Notably, miR-300 may suppress epithelial–mesenchymal transition (EMT) by increasing the expression levels of E-cadherin and decreasing the expression levels of N-cadherin, Vimentin, Snail1 and MMP-2. Moreover, miR-300 mRNA levels were altered due to the differential expression of ET-1. In conclusion, the results of the present study demonstrate that miR-300 may act as a tumor suppressor by inhibiting EMT, suggesting that miR-300 may be a potential biomarker and anticancer therapeutic target for OSCC.
Materials and methods
Tissue samples
A total of 120 specimens of OSCC were obtained from patients who underwent surgical resection in the Department of Oral and Maxillofacial Surgery, the School of Stomatology, China Medical University between 2008 and 2012. These tumor specimens were stored at −80°C. Experiments using human samples were approved by the ethics committee of the School of Stomatology, China Medical University, and written informed consent was obtained from the donors.
Cell culture
Human OSCC cell lines Tca8113, Cal-27 and HaCat cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The Tca8113 cells were maintained in 1640 medium and Cal-27 and HaCat cells were maintained in DMEM medium; 10% of fetal bovine serum and 1% of penicillin–streptomycin solution were added to them. All the cell lines were maintained at 37°C in the presence of 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the frozen tissues and cells using TRIzol (Takara Corporation, Dalian, China) according to the manufacturer’s instructions. Using standard spectrophotometric methods, RNA concentration was quantified and its purity was determined. RNA (1 μg) was reverse transcribed using Harrpin-it™ microRNA and U6 snRNA Normalization RT-PCR Quantitation Kit (GenePharma Co, Shanghai, China). The RT reaction system was as follows: 5× MMLV RT buffer 4 μL, dNTP 0.75 μL, miRNA and U6 snRNA RT primer mix (1 μM) 1.2 μL, MML Reverse Transcriptase (200 U/μL)2 0.2 μL, RNA 1 μg; RNase-free H2O was added up to 20 μL. The following thermal cycling conditions were employed: 25°C for 30 minutes, 42°C for 30 minutes and 85°C for 5 minutes. qPCR reaction system: 2× Real-time PCR Master Mix (SYBR) 10 μL, miRNA and U6 snRNA-specific primer set (10 μM)1 0.4 μL, ROX reference dye (50×)3 0.4 μL, Taq DNA polymerase (5 U/μL) 0.2 μL, miRNA RT product 2 μL; sterilized H2O was added up to 20 μL. The cycling conditions were as follows: 95°C for 3 minutes, 95°C for 12 seconds and 62°C for 40 seconds.
To examine the expression of E-cadherin, N-cadherin, Vimentin, Snail1 and MMP-2, cDNA served as the template for amplification of PCR using a cDNA Synthesis kit (PrimeScript™ RT reagent kit, Takara), according to the manufacturer’s instructions. The RT reaction system was as follows: 5× PrimeScript RT Master Mix 4 μL, RNA 1 μg; RNase-free distilled H2O was added up to 20 μL. The following thermal cycling conditions were employed: 37°C for 15 minutes and 85°C for 5 minutes. qPCR was conducted using the SYBR Premix Ex Taq II kit (Takara Corporation) with a qPCR reaction system: SYBR Premix Ex Taq II 10 μL, cDNA 1 μL, forward primer 0.5 μL, reverse primer 0.5 μL and sterile water 8 μL. The cycling conditions were as follows: 95°C for 1 minute, 94°C for 30 seconds, 58°C for 30 seconds and 72°C for 10 seconds.
Gene expression was normalized to GAPDH as an internal control, and the relative level of miR-300 was calculated using the 2−ΔΔCt method.
Western blotting
Protein concentrations were determined by the bicinchoninic acid method. Next, samples corresponding to 50 μg of total protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 10% gel and then transferred onto polyvinylidene difluoride membranes (Sigma, Shanghai, China). After blockage with 5% non-fat milk, the membrane was probed with primary antibodies including E-cadherin (1:1,000, Cell Signaling Technology), N-cadherin (1:500, Cell Signaling Technology), Vimentin (1:1,000, Cell Signaling Technology), Snail1 (1:1,000, Cell Signaling Technology), MMP-2 (1:1,000, Cell Signaling Technology) and GAPDH antibodies (1:2,000, Santa Cruz Biotechnology Inc.) and then probed with secondary antibodies (1:5,000, Santa Cruz Biotechnology Inc.). Proteins were visualized by means of Thermo Pierce ECL (Thermo Fisher Scientific, Waltham, MA, USA).
Cell transfection
MiR-300 mimics, miR-300 inhibitor and control were synthesized by GenePharma Co. (GenePharma Co, Shanghai, China). The cells were seeded in six-well plates and were transfected using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) following the manufacturer’s protocol.
- miR-300 mimics sense: 5′-UAUACAAGGGCAGACUCUCUCU-3′, antisense: 5′-AGAGAGUCUGCCCUUGUAUAUU-3′
- negative control sense: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′-ACGUGACACGUUCGGAGAATT-3′
- miR-300 inhibitor: 5′-AGAGAGAGUCUGCCUUGUAUA-3′
- miR inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-etrazolium, inner salt (MTS) assay
Proliferation assays were conducted using the Cell Proliferation Assay kit (Promega, Madison, WI, USA). OSCC cells were plated in 96-well plates (2×103 cells/well) and cultured at 37°C for 18 hours prior to transfection. Subsequently, the cells were transfected with miR-300 mimics and miR-NC, in addition to miR-300 inhibitor and miR-inhibitor NC. The culture plates were taken out at different time periods (24, 48 and 72 hours) and continuously cultured for 2 hours after the addition of 10 μL MTS in each well. The OD value was measured at a wavelength of 570 nm using enzyme-linked immunosorbent assay.
Transwell assays
OSCC cells were transfected for 48 hours. The cells (5×104 cells/well) were resuspended in serum-free medium and plated onto the upper chamber (Corning Costar, Tewksbury, MA, USA). Then, 0.5 mL of medium containing 10% fetal bovine serum (Clark, Beijing, China) was added in the lower chamber to act as chemoattractant. Transwell chambers coated with Matrigel (Corning Inc.) were used. Cells in the Transwell plates were incubated at 37°C in a humidified atmosphere with 5% CO2 for 48 hours. The adherent cells on the upper surface of the insert membrane were carefully removed with cotton tips. Cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 15 minutes, permeabilized in 0.1% Triton X-100 and stained with 0.1% crystal violet for 30 minutes. The invaded cells were imaged and quantified using a microscope (Olympus, Beijing, China) in five separate fields per membrane.
Statistical analysis
Statistical analysis was carried out in the IBM SPSS 21.0 software (IBM Corporation, Armonk, NY, USA). Student’s t-test was used to examine statistical differences between two groups. Correlations between the miR-300 expression and clinical characteristics of OSCC patients were assessed by chi-squared test. In addition, overall survival (OS) and disease-free survival (DFS) were calculated by the Kaplan–Meier method. To determine the effect of particular prognostic factors on survival, a multivariate analysis was performed according to the Cox regression model. P<0.05 was assumed to indicate a statistically significant difference.
Results
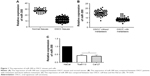
Downregulation of miR-300 in OSCC
In order to examine the oncogenic role of the miR-300, we compared the miRNA expression between the OSCC tissues and normal oral mucosa tissues. qRT-PCR showed that compared with healthy normal tissues, the expression of miR-300 was obviously reduced in OSCC tissues (P<0.05; Figure 1A). Then, we divided the OSCC patients into two groups: patients with metastasis and those without metastasis. Compared with patients without metastasis, patients with metastasis showed significantly decreased level of miR-300 (P<0.05; Figure 1B). Furthermore, compared with HaCat cells, the expression of miR-300 in Tca8113 and Cal-27 cell lines was significantly decreased (P<0.05; Figure 1C).
The relationship between miR-300 and the clinicopathologic characteristics of the OSCC patients is summarized in Table 1. Correlations between miR-300 and gender, age and tumor site were not statistically significant. However, the results showed that miR-300 was closely related to TNM classification (P<0.05; Table 1).
  | Table 1 The relationship between miR-300 and the clinicopathologic characteristics in OSCC patients |
To elucidate the prognostic role of miR-300, we examined the relationship between miR-300 expression and patient outcome with long-term follow-up. Kaplan–Meier analysis further showed patients with relatively lower level of miR-300 had significantly decreased OS (P<0.0001; Figure 2A) and DFS (P<0.0001; Figure 2B). Univariate and multivariate analyses were carried out using Cox proportional hazard model to evaluate the impact of miR-300 expression and clinicopathologic factors on the prognosis of OSCC patients. As shown in Table 2, univariate Cox regression analysis suggested that differentiation (P=0.031 for OS, P=0.045 for DFS), TNM stage (P=0.041 for OS, P=0.035 for DFS), miR-300 expression (P=0.028 for OS, P=0.025 for DFS) as well as tobacco usage (P=0.033 for OS, P=0.040 for DFS) were significantly associated with poor OS and DFS. Multivariate analyses showed TNM stage (P=0.049 for OS, P=0.043 for DFS), miR-300 expression (P=0.035 for OS, P=0.030 for DFS) as well as tobacco usage (P=0.045 for OS, P=0.049 for DFS) were associated with poor OS and DFS.
miR-300 inhibits proliferation and invasion of OSCC cells
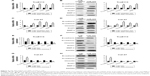
In order to analyze the association between miR-300 and OSCC, OSCC cells were transfected with miR-300 mimics or miR-300 inhibitor and the transfection efficiency was detected by RT-qPCR (Figure 3A and C). Overexpression of miR-300 significantly inhibited cell proliferation (Figure 3B), On the contrary, miR-300 inhibitor significantly promoted cell proliferation (Figure 3D). Overexpression of miR-300 significantly inhibited cell invasion (Figure 3E) and miR-300 inhibitor promoted cell invasion (Figure 3F) of Tca8113 and Cal-27 cell lines.
miR-300 inhibits the EMT of OSCC cells
To explore the molecular mechanism of miR-300 in OSCC, we assessed the role of miR-300 in the EMT of OSCC cells. We performed Western blot and RT-qPCR to detect the expression of E-cadherin, N-cadherin, Vimentin, Snail1 and MMP-2 in the OSCC cells transfected with miR-300 mimics and inhibitor. The results of RT-qPCR (Figure 4A) and Western blot (Figure 4B) demonstrated that overexpression of miR-300 in Tca8113 cells increased the expression of E-cadherin and decreased N-cadherin, Vimentin, Snail1 and MMP-2 expression significantly. The results of RT-qPCR (Figure 4C) and Western blot (Figure 4D) in Cal-27 cells is the same. On the other hand, RT-qPCR (Figure 4E) and Western blot (Figure 4F) showed that miR-300 inhibitor in Tca8113 cells reduced E-cadherin expression and increased N-cadherin, Vimentin, Snail1 and MMP-2 expression significantly. The results of RT-qPCR (Figure 4G) and Western blot (Figure 4H) in Cal-27 cells is the same.
Endothelin-1 (ET-1) inhibiting miR-300 in OSCC
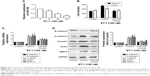
The results of cell proliferation and invasion in this study indicated that miR-300 may inhibit the proliferation and invasion OSCC cells. To further understand the mechanisms of miR-300 in OSCC metastasis, ET-1 was used. We found that miR-300 expression was decreased in a dose-dependent manner after ET-1 treatment (Figure 5A).
When we transfected OSCC with an miR-300 mimic and then treated them with ET-1, we confirmed the role of miR-300 in cell invasion. The data indicate that the miR-300 mimic inhibited ET-1–induced invasion (Figure 5B). We also find that the miR-300 mimic but not the control miRNA abolished ET-1–induced EMT (Figure 5C and D).
Discussion
In OSCC patients, metastasis is the primary cause of mortality. Thus, it is very important to understand the molecular mechanisms of metastasis. Recently, miRNAs have been reported to promote or suppress tumor metastasis,13,14 providing a new perspective on the metastatic process. Emerging research suggests that miRNAs play essential roles in the progression of OSCC.15–19 Nonetheless, research on the role of miRNAs in OSCC metastasis is lacking. EMT, which enables epithelial cells to acquire invasive mesenchymal phenotype, is attracting increasing attention as an important mechanism for the initial step of metastasis.20 In this study, we observed that miR-300 played an important role as a suppressor of EMT in OSCC, which, in turn, inhibited metastasis.
To gain further insights into the role of miR-300 in OSCC metastasis, the expression of miR-300 was detected in 120 OSCC samples. We found that miR-300 expression was significantly decreased in OSCC tissue. Furthermore, significantly lower levels of miR-300 were found in patients with metastasis than those in patients without metastasis. Decreased expression of miR-300 was associated with adverse clinicopathologic features and poor prognosis of OSCC patients. Functionally, miR-300 was found to inhibit the proliferation and invasion of OSCC cells. Moreover, through inhibiting EMT phenotype (as suggested by increased E-cadherin and decreased N-cadherin, Vimentin, Snail and MMP-2), we further explored whether miR-300 inhibited the metastasis of OSCC cells, and the data of RT-qPCR and Western blot showed that miR-300 could inhibit the EMT of OSCC cells.
Considerable evidence suggests that tumor cells expressing aberrant levels of ET-1 facilitate tumor development and progression. However, the molecular mechanisms of ET-1–regulated EMT by miRNA in human oral cancers are not well characterized. Our study indicates that miR-300 is downregulated in response to ET-1, and that transfection of cells with miR-300 mimic reduces ET-1–induced cell invasion. The ET-1 signaling has a distinct function in OSCC, namely, regulation of EMT and cell invasion by repressing miR-300.
However, there are some limitations of our study. First, the sample size in the subgroup analysis is not big enough; thus, the statistical power is limited. Second, the relevant mechanisms have not been identified. In our future work, we plan to do more in-depth research and analysis. Furthermore, animal experiments need to be devised to investigate the effects of miR-300 on tumor growth in vitro and in vivo.
In conclusion, the key finding in our study is that miR-300 suppresses metastasis of OSCC by inhibiting EMT. These findings indicate that high miR-300 expression may be a useful therapeutic target for OSCC.
Acknowledgment
This study was supported by the Natural Science Foundation of Liaoning Province (20170541012).
Disclosure
The authors report no conflicts of interest in this work.
References
Liao L, Wang J, Ouyang S, Zhang P, Wang J, Zhang M. Expression and clinical significance of microRNA-1246 in human oral squamous cell carcinoma. Med Sci Monit. 2015;21:776–781. | ||
Shekhar S, Angadi PV. Evaluation of paxillin expression in patients with oral squamous cell carcinoma: an immunohistochemical study. J Oral Maxillofac Pathol. 2017;21(2):318–319. | ||
Rusek AM, Abba M, Eljaszewicz A, Moniuszko M, Niklinski J, Allgayer H. MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol Cancer. 2015;14(1):34. | ||
Bai X, Meng L, Sun H, Li Z, Zhang X, Hua S. MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting Runx2. Cell Physiol Biochem. 2017;43(2):757–767. | ||
Guan SS, Chang J, Cheng CC, et al. Afatinib and its encapsulated polymeric micelles inhibits HER2-overexpressed colorectal tumor cell growth in vitro and in vivo. Oncotarget. 2014;5(13):4868–4880. | ||
Luo BH, Xiong F, Wang JP, et al. Epidermal growth factor-like domain-containing protein 7 (EGFL7) enhances EGF receptor-AKT signaling, epithelial–mesenchymal transition, and metastasis of gastric cancer cells. PLoS One. 2014;9(6):e99922. | ||
Vaiopoulos AG, Kostakis ID, Gkioka E, et al. Detection of circulating tumor cells in colorectal and gastric cancer using a multiplex PCR assay. Anticancer Res. 2014;34(6):3083–3092. | ||
Sun L, Liu L, Fu H, Wang Q, Shi Y. Association of decreased expression of serum miR-9 with poor prognosis of oral squamous cell carcinoma patients. Med Sci Monit. 2016;22:289–294. | ||
Shen Z, Li C, Zhang K, et al. The up-regulation of miR-300 in gastric cancer and its effects on cells malignancy. Int J Clin Exp Med. 2015;8(5):6773–6783. | ||
Ge W, Han C, Wang J, Zhang Y. MiR-300 suppresses laryngeal squamous cell carcinoma proliferation and metastasis by targeting ROS1. Am J Transl Res. 2016;8(9):3903–3911. | ||
He FY, Liu HJ, Guo Q, Sheng JL. Reduced miR-300 expression predicts poor prognosis in patients with laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(4):760–764. | ||
Xu XH, Li DW, Feng H, Chen HM, Song YQ. MiR-300 regulate the malignancy of breast cancer by targeting p53. Int J Clin Exp Med. 2015;8(5):6957–6966. | ||
Rasul A, Khan M, Yu B, et al. Isoalantolactone, a sesquiterpene lactone, induces apoptosis in SGC-7901 cells via mitochondrial and phosphatidylinositol 3-kinase/Akt signaling pathways. Arch Pharm Res. 2013;36(10):1262–1269. | ||
Hong KJ, Wu DC, Cheng KH, Chen LT, Hung WC. RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation. J Cell Physiol. 2014;229(2):191–201. | ||
Yu T, Liu K, Wu Y, et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene. 2014;33(42):5017–5027. | ||
Jia LF, Huang YP, Zheng YF, et al. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50(11):1062–1071. | ||
Ren Y, Zhu H, Chi C, Yang F, Xu X. MiRNA-139 regulates oral cancer Tca8113 cells apoptosis through Akt signaling pathway. Int J Clin Exp Pathol. 2015;8(5):4588–4594. | ||
Chang KW, Kao SY, Wu YH, et al. Passenger strand miRNA miR-31* regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013;49(1):27–33. | ||
Chawla JP, Iyer N, Soodan KS, Sharma A, Khurana SK, Priyadarshni P. Role of miRNA in cancer diagnosis, prognosis, therapy and regulation of its expression by Epstein–Barr virus and human papillomaviruses: with special reference to oral cancer. Oral Oncol. 2015;51(8):731–737. | ||
Pecinaslaus N, Cicvarapecina T, Kafka A. Epithelial-to-mesenchymal transition: possible role in meningiomas. Front Biosci (Elite Ed). 2011;4(3):889–896. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.