Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-21-5p promotes proliferation of gastric cancer cells through targeting SMAD7
Authors Jiang Y, Zhang M, Guo T, Yang C, Zhang C, Hao J
Received 25 January 2018
Accepted for publication 20 May 2018
Published 15 August 2018 Volume 2018:11 Pages 4901—4911
DOI https://doi.org/10.2147/OTT.S163771
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Yinan Jiang,1 Meiling Zhang,2 Tangxi Guo,1 Chaogang Yang,1 Chunxiao Zhang,1 Jinjin Hao2
1Department of Gastrointestinal Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, China; 2Department of Pediatrics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China
Background: MicroRNAs could target multiple genes by regulating the translation or degradation of mRNAs, and are involved in functions such as signal transduction pathways affecting the physiological functions of normal or tumor cells.
Methods: In this study, the expressions of miRNA-21-5p in gastric cancer tissues and SGC-7901 cells were analyzed, and the effects of miRNA-21-5p on cell proliferation, migration, invasion, and apoptosis and the expressions of target proteins SMADs in SGC-7901 cells were evaluated. Inflammatory factors interleukin 1β and tumor necrosis factor α in siRNA-transfected SGC-7901 cells were determined by enzyme-linked immunosorbent assay.
Results: MiRNA-21-5p was consistently upregulated in gastric cancer tissues and SGC-7901 cells compared to normal tissues or cells. The knockdown of miRNA-21-5p with antisense oligonucleotides suppressed cell proliferation, migration, and invasion as well as inflammatory response, and promoted cell apoptosis and SMAD7 expression.
Conclusion: These results indicate SMAD7 may mediate the oncogenic properties of miRNA-21-5p in gastric cancer, and miRNA-21-5p would be a promising strategy for the treatment of gastric cancer.
Keywords: miRNA-21-5p, gastric cancer, SMAD7, SGC-7901, migration, invasion
Introduction
Gastric cancer (GC) is the fourth most common malignancy and the third leading cause of cancer-related death worldwide in 2015.1 The incidence and mortality of GC are the highest in East Asia (China, Korea, and Japan), and it is the second most lethal cancer in China.2 However, owing to few diagnostic methods for early detection of GC, most GC patients are diagnosed at middle or late stage, with 35% and 90% of GC patients having distant metastases and lymph node metastases, respectively.3 Thus, there is an urgent search for new diagnosis and treatment methods for GC. Now, epigenetic changes including microRNAs (miRNAs) and long non-coding RNAs have been confirmed to be involved in the pathogenesis of GC.4
MiRNAs, as small non-coding RNA molecules (20–22 nucleotides long), play significant roles in post-transcriptional regulation, such as silencing target mRNAs by binding to miRNA recognition elements in the 3′ untranslated region (3′UTR) and thus decreasing their protein levels.5,6 MiRNAs as post-transcriptional regulators of gene expression are critical in the regulation of various bioprocesses, including cell differentiation, proliferation, and apoptosis.7 Accumulating evidence indicates miRNA mutations or mis-expression correlate with various human cancers and some miRNAs may function as oncogenes or tumor suppressors in normal and tumor tissues, including GC.8,9 Studies describing the altered expressions of miRNAs in cancer tissues compared to normal tissues suggest that miRNAs are potentially novel clinical and prognostic biomarkers for the detection of various cancers.10,11 The discovery of miRNAs and their expression profiles among different diseases indicates the common miRNA-21 could serve as a potential biomarker for GC detection.12 MiRNA-21-5p (miR-21-5p; hsa-mir-21-5p) located on 17q23.1 has been identified in several tumors and is in particular highly expressed in GC tissues, which indicate an enhanced expression with the disease advancement.
As described earlier, miR-21-5p seems to play an important oncogenic role in GC. In this study, we aimed to elucidate the effects of miR-21-5p on GC and to investigate its underlying mechanisms.
Materials and methods
Patients and tissue samples
Twenty-five GC tissue samples were obtained from 25 GC patients (13 males, 12 females; 34–76 years old, mean age 55.9 years, SD 11.4 years) histologically diagnosed between March 2014 and January 2016 at Zhongnan Hospital of Wuhan University (Wuhan, China). None of the patients received chemotherapy or radiotherapy. At the same time, 25 cases of normal tissues (13 males, 12 females, mean age 52.6 years, SD 13.4 years, range 27–73 years) were purchased from Zhongnan Hospital of Wuhan University. All subjects agreed to be included, and provided written informed consent; the study was conducted in accordance with the Declaration of Helsinki. The protocol was approved by Zhongnan Hospital of Wuhan University.
Cell culture
GC cell lines SGC-7901, BGC-823, MGC-803, and normal gastric epithelial cell line GES-1 were purchased from Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). All cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin, and incubated at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNAs of tissues or cell lines were extracted using TRIzol reagent according to manufacturer’s protocol. The cDNA was transcribed using a Prime Script™ RT Master Mixture according to the manufacturer’s protocol (Takara Biotechnology, Daliang, China). qPCR was performed on an ABI Prism PCR system and the expression of miR-21-5p was quantified via the SYBR Green I fluorescent dye method (Takara Biotechnology). PCR was initiated at 95°C for 2 min, followed by 36 cycles at 95°C for 20 s and 58°C for 25 s. The following forward and reverse primers were used: 5′-GGGGTAGCTTAT CAGACTGATG-3′, 5′-TGTCGTGGAGCGGCAATTG-3′ (miR-21-5p); 5′-CGCTTCGGCACATATACTA-3′, 5′-CGCTTCACGAATTTGCGTGTCA-3′ (U6). The relative level of miR-21-5p was calculated with the comparative 2−ΔΔCt method normalized by U6.
Immunohistochemistry (IHC)
Tissues were processed by routine dehydration, transparency, paraffin leaching, and paraffin embedding. Slices were washed with phosphate buffer solution (PBS) twice and added with 5% bovine serum albumin sealing liquid. Antibody (rabbit anti-rabbit SMAD7) was added at 37°C for 1 h, followed by washing with PBS twice. The labeled antibody was added at 37°C for 30 min, followed by washing with PBS. The diaminobenzidine (DAB) dye was added at room temperature, and the reaction time of 5 min was controlled by observation under a microscope, followed by washing with distilled water. Then the samples were stained slightly with hematoxylin and eosin. Results were observed by microscopy.
The target gene SMAD7 of miR-21-5p predicted by Targetscan
Targets of miR-21-5p were searched on Targetscan (www.targetscan.org/vert_71/), and the results were further confirmed by PicTar29 (pictar.mdc-berlin.de/) and MicroRNA.org30 (www.microrna.org), suggesting SMAD7 is a potential target of miR-21-5p. To further confirm that SMAD7 is directly targeted by miR-21-5p, we obtained more information about the 3′UTR of SMAD7 mRNAs on Targetscan.
MiR-21-5p transfection and luciferase reporter assay
The sequence of SMAD7 was searched in the gene bank of US National Center for Biotechnology Information (NCBI). The 3′UTR of SMAD7 was amplified and cloned into the specific vector of SMAD7-UTR-pISo. Luciferase reporter plasmids of WT-SMAD7 mRNA and MUT-SMAD7 mRNA were constructed using the clones. The SGC-7901 cells were cultured for 24 h, while SMAD7-UTR-pISo plasmid and miR-21-5p inhibitors or Mu-SMAD7-UTR-pIS0 plasmid and miR-21-5p inhibitors were co-transfected into cells using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Relative luciferase activity of SMAD7 was detected with Dual-Luciferase Reporter Assay.
Cell proliferation analysis
The proliferation of GES-1, SGC-7901, BGC-823, and MGC-803 cells was detected by Cell Counting kit 8 in 96-well plates according to the manufacturer’s protocols. About 3×103 cells were seeded into wells; transfected with miR-21-5p mimics, inhibitors, and negative miRNA; and grew for 24 h at 37°C. After medium was changed, transfection cells were treated for the next experiment.
Wound healing assay
SGC-7901 cells transfected with miR-21-5p mimics, inhibitors, and negative miRNA were seeded in six-well plates when the fusion reached above 90%. A wound was mimicked by making a scratch at the middle of the well using a 100 μL of tip, and washed with PBS twice. Cells in the six-well plates were cultured with DMEM without serum for 24 h. The healing of the wound was observed under an inverted microscope, and the migrating distance of the cell monolayer to near the wounded area during this time period was measured.
Transwell assay
SGC-7901 cells were re-suspended with DMEM without serum at a concentration of 1×105/mL, and then the cells (200 μL) were seeded onto the upper well of matrigel-coated (Sigma Aldrich Co., St Louis, MO, USA) 8 μm pore transwell inserts (Sigma Aldrich Co.). The lower chamber was added with DMEM containing 10% FBS. After 24 h of incubation, cells in the chamber were removed with a cotton swab. After fixed staining with 0.1% crystal violet, random pictures were taken and cells were counted under the microscope.
Apoptosis analysis by Hoechst 33258 staining
SGC-7901 cells transfected with miR-21-5p mimics, inhibitors, and negative miRNA were grown for 24 h. The cells were centrifuged at 1,000 rpm for 5 min and washed with PBS. Then the cells were stained with Hoechst 33258 (50 μg/mL; Boster Biotechnology Company, Wuhan China) and incubated at 37°C for 30 min in dark. After that, the cells were washed, resuspended with PBS, observed under a fluorescence microscope (×400; Nikon, Tokyo, Japan), and analyzed on DP2-BSW. The apoptotic cells shrunk with condensed chromatin.
Cytokine analysis
SGC-7901 cells in six-well plates were transfected with miR-21-5p mimics, miR-21-5p inhibitors, and negative miRNA for 24 h. The concentrations of interleukin (IL)-1β and tumor necrosis factor (TNF)-α in the supernatants of transfected cells were analyzed by enzyme-linked immunosorbent assay (ELISA; RD Systems, Minneapolis, MN, USA) according to the manufacturer’s instruction. Cytokine concentrations were calculated by standard curves.
Immunoblotting
Cell lysates were determined by a bicinchoninic acid (BCA) protein assay kit. The proteins were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After that, the gels were transferred to polyvinylidene difluoride (PVDF) membranes, which were incubated with rabbit polyclonal anti-SMAD2 (dilution 1:800), anti-SMAD3 (1:1,000), anti-SMAD7 (1:1,200), and anti-GAPDH (1:600) antibodies (all BD, Franklin Lakes, NJ, USA) overnight at 4°C. After washing with PBS three times, the membranes were incubated with a secondary polyclonal peroxidase labeled antibody for 2 h, and detected using autoradiography films (Amersham HyperFilm ECL; GE Healthcare, Fairfield, Connecticut, USA). Quantification was performed on Quantity One.
Statistical analysis
All statistical analyses were conducted on SPSS 16.0 with the significance level at P<0.05. All experiments were repeated three times and a representative experiment result was shown with SD.
Results
Analysis of miR-21-5p in GC tissues or cells
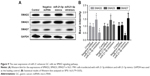
MiR-21-5p is abnormally expressed in digestive cancers. Here the expression levels of miR-21-5p in GC tissues (Figure 1A) and SGC-7901 cells (Figure 1B) were determined by qRT-PCR. Clearly, miR-21-5p expressions were upregulated significantly in both GC cells and SGC-7901 cells compared with normal tissues or cells.
Analysis of target genes of miRNA-21-5p by Targetscan
Since miRNAs play a gene silencing function by binding target genes at post-transcriptional and transcriptional levels, we predicted the target genes of miRNA-21-5p on Targetscan. Interestingly, we found about 21 target genes by bioinformatic analysis and were more interested in SMAD7 (Figure 2A). We acquired the information about miRNA-21-5p binding with the sites in the 3′UTR of SMAD7 (Figure 2B).
Analysis of SMAD7 expression in GC tissues or cells
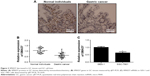
The 3′UTR of SMAD7 gene was a target site of miR-21-5p. SMAD7 proteins in GC tissues were detected by IHC (Figure 3A). Additionally, the expression levels of SMAD7 genes in GC tissues (Figure 3B) and SGC-7901 cells (Figure 3C) were determined by qRT-PCR. Experiments indicated SMAD7 genes or proteins were decreased in tissues and SGC-7901 cells compared to normal tissues or cells.
SMAD7 as a target gene of miR-21-5p
Targetscan predicted that miR-21-5p bound the 3′UTR of SMAD7 (Figure 4A). The relationship between miR-21-5p and SMAD7 was validated by luciferase reporter assay. Plasmids of SMAD7-UTR-pISo (WT) and Mu-SMAD7-UTR-pIS0 (MUT) were constructed and transfected into SGC-7901 cells. Results showed the luciferase activity of SMAD7-UTR-pISo (WT) was significantly enhanced by miR-21-5p inhibitors, and significantly weakened by SGC-7901 cells transfected with miR-21-5p mimics (Figure 4B). The miR-21-5p expression of SGC-7901 cells was downregulated evidently after transfection with miR-21-5p inhibitors, but was remarkably upregulated after transfection with miR-21-5p mimics (Figure 4C). Meanwhile, the SMAD7 expression of SGC-7901 cells was significantly upregulated by transfection with miR-21-5p inhibitors, but was obviously downregulated by transfection with miR-21-5p mimics (Figure 4D). In all, SMAD7 is targeted and regulated by miR-21-5p.
The knockdown of miR-21-5p inhibited GC cell growth
It is well established that miR-21-5p expression promotes the proliferation and invasiveness of breast cancer cells and prostate cancer cells. Thus, we investigated if GC cell growth could be inhibited in response to miR-21-5p knockdown. The proliferation levels of GES-1, SGC-7901, BGC-823, and MGC-803 cells (Figure 5A–D, respectively) transfected with miR-21-5p inhibitors, miR-21-5p mimics, and negative miRNAs were analyzed by CCK-8 assay, with GES-1 cells as the control. As expected, miR-21-5p knockdown dramatically inhibited the proliferation levels of SGC-7901, BGC-823, and MGC-803 cells, while the over-expression of miR-21-5p induced an increase of cell proliferation in these three GC cell lines.
MiR-21-5p knockdown decreased the migration and invasion of SGC-7901 cells
With invasion and metastasis abilities, cancer cells can pass through the basement membrane, migrate to distant tissues, and invade other tissues. In this study, the effects of miR-21-5p on cell migration and invasion were evaluated. The wound-healing assay revealed miR-21-5p knockdown significantly decelerated the scratch closure rate of SGC-7901 cells compared with the control (Figure 6A), suggesting the mobility of SGC-7901 cells was suppressed. Furthermore, the invasion assay demonstrated that the down-expression of miR-21-5p significantly inhibited the invasion capacity of SGC-7901 cells compared with the control (Figure 6B). These results suggest miR-21-5p knockdown could suppress the migration and invasion of GC cells.
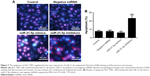
Apoptosis analysis of GC cells silenced by siRNA
To investigate the effect of miR-21-5p on GC cells, we transfected SGC-7901 cells with miR-21-5p mimics, inhibitors, and negative miRNA for 24 h, followed by staining with Hoechst 33258 for apoptosis analysis (Figure 7A). Results suggest the knockdown of miR-21-5p significantly promoted the apoptosis of SGC-7901 cells compared with the control, but the over-expression of miR-21-5p inhibited the apoptosis of SGC-7901 cells, indicating miR-21-5p promoted the growth of SGC-7901 cells evaluated by the statistical data of apoptosis (Figure 7B).
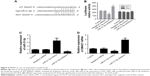
Knockdown of miR-21-5p downregulates the expression of IL-1β and TNF-α
Since the transforming growth factor (TGF)-β1/Smads signaling pathway is closely related to inflammation regulation, the expression levels of inflammatory cytokines IL-1β and TNF-α were determined to study the functions of SMAD7 regulated by miR-21-5p (Figure 8). As expected, the expressions of IL-1β and TNF-α were significantly downregulated in SGC-7901 cells transfected with miR-21-5p inhibitors, but were significantly upregulated after transfection with miR-21-5p mimics compared with the control. These results suggest miR-21-5p may negatively regulate the expressions of tumor-associated cytokines by targeting SMAD7 in SGC-7901 cells.
The over-expression of miR-21-5p increases SMAD protein in GC cells
As miR-21-5p was enriched in GC tissues and cells, the ability to modulate the SMAD protein expressions in SGC-7901 cells regulated by siRNAs of miR-21-5p was assessed by Western blot (Figure 9). Results show SMAD7 proteins were increased significantly in SGC-7901 cells transfected with miRNA-21-5p inhibitors, but the expression of SMAD7 was inhibited significantly by the over-expression of miRNA-21-5p. Meanwhile, SMAD2 proteins of SGC-7901 cells decreased significantly on transfection with miR-21-5p inhibitors, but were increased obviously by the over-expression of miRNA-21-5p, indicating miR-21-5p functions with an opposite role in the expressions of SMAD proteins.
Discussion
Increasing evidence has shown that miRNAs play significant functions in the carcinogenic process,13,14 and reports show more than 50% of miRNAs are located in cancer-associated genomic regions or in fragile sites.27 MiRNA profiles are significantly associated with essential tumor features and patient survival. Investigation into the expression patterns and biological functions of miRNAs in GC suggests miRNAs are potential novel therapeutic strategies for GC in future. MiR-21-5p as an important oncogenic miRNA was recently reported to be highly upregulated in multiple tumors and the regulatory mechanisms were well characterized.15–17 MiR-21-5p could promote tumor growth and metastasis by targeting several tumor suppressors, including PTEN, PDCD4, Bcl2, and TIMP3 in breast cancer, suggesting that miR-21-5p might be involved in regulating angiogenesis.18–20 Moreover, miR-21 plays important roles in the proliferation and migration of GC cells. Here we found miRNA-21-5p increased significantly in GC tissues and cell lines. Interestingly, predictions on Targetscan identified 21 target genes of miRNA-21-5p. Particularly, we were interested in SMAD7 gene and did thorough experiments in SGC-7901 cells.
SMAD7 is a member of the SMAD family, which belongs to TGF-β superfamily of ligands and is involved in cell signaling. The over-expression of miR-21 promotes the TGF-β pathway by silencing SMAD7, which initiates epithelial–mesenchymal transformation and invasion.21 Our results showed SMAD7 levels decreased in GC tissues and GC cell lines. Additionally, the knockdown of miRNA-21-5p remarkably decreased the proliferative capacity and obviously promoted the apoptosis of SGC-7901 cells. MiR-21 acts as an important repressor of SMAD-dependent TGF-β pathway by targeting PTEN and SMAD7, while the activation of miR-21 expression can be mediated by a TGF-β activity.22–24 SMAD7 could block TGF-β signaling by preventing formation of Smad2/Smad4 complexes which initiate the TGF-β signaling.25 In addition, the TGF-β1/SMADs signaling pathway is reportedly involved in inflammation regulation. In SMAD7 knockout mice with diabetic and obstructive nephropathies, a severe renal inflammatory reaction could be induced, in which IL-1, TNF-α, and MCP-1 levels increased significantly, suggesting SMAD7 is able to inhibit the inflammatory response.26,27
In this study, miR-21-5p may negatively regulate the expression of tumor-associated cytokines IL-1β and TNF-α by targeting SMAD7 in SGC-7901 cells. Additionally, the over-expression of miRNA-21-5p increased the SMAD7 protein expression and inhibited the SMAD2/SMAD3 expressions, suggesting SMAD7 proteins were regulated by miRNA-21-5p and SMAD2/SMAD3 could be negatively modulated by SMAD7. Overall, these results broaden our understanding about the mechanism of miR-21-5p in mediating SMAD7, and provide novel prognostic and predictive factors for GC treatment and the design of novel miRNA-based anticancer therapeutic strategies. These results also support proceeding to additional preclinical studies concerning miR-21-5p inhibitors or mimics as therapeutic strategies to suppress or promote the proliferation, apoptosis, migration, and invasion in vivo.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;523(7517):202–209. | ||
Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11(2):136–146. | ||
Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. | ||
Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev Mol Diagn. 2007;7(5):569–603. | ||
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. | ||
Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Molecular cancer. 2006;5:24. | ||
Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9(3):189–198. | ||
Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Inves. 2008;88(12):1358–1366. | ||
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. | ||
Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. | ||
Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3(3):251–255. | ||
Li S, Liang Z, Xu L, et al. MicroRNA-21: a ubiquitously expressed pro-survival factor in cancer and other diseases. Mol Cell Biochem. 2012;360(1–2):147–158. | ||
Bonci D. MicroRNA-21 as therapeutic target in cancer and cardiovascular disease. Recent Pat Cardiovasc Drug Discov. 2010;5(3):156–161. | ||
Yang Y, Chaerkady R, Beer MA, et al. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009;9(5):1374–1384. | ||
Wickramasinghe NS, Manavalan TT, Dougherty SM, et al. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37(8):2584–2595. | ||
Song B, Wang C, Liu J, et al. Micro-RNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 2010;29:29. | ||
Chen L, Li Y, Fu Y, et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PLoS One. 2013;8(1):e54213. | ||
Ardite E, Perdiguero E, Vidal B, et al. PAI-1-regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J Cell Biol. 2012;196(1):163–175. | ||
Zhong X, Chung AC, Chen HY, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56(3):663–674. | ||
McClelland AD, Herman-Edelstein M, Komers R, et al. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin Sci (Lond). 2015;129(2):1237–1249. | ||
Marts LT, Green DE, Mills ST, et al. MiR-21-mediated suppression of Smad7 induces TGFβ1 and can be inhibited by activation of Nrf2 in alcohol-treated lung fibroblasts. Alcohol Clin Exp Res. 2017;41(11):1875–1885. | ||
Chen HY, Huang XR, Wang WS, et al. The protective role of Smad7 in diabetic kidney disease: mechanism and therapeutic potential. Diabetes. 2011;60(2):590–601. | ||
Huang XR, Zhou LHR. Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol Dial Transplant. 2009;24(5):1443–1454. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.